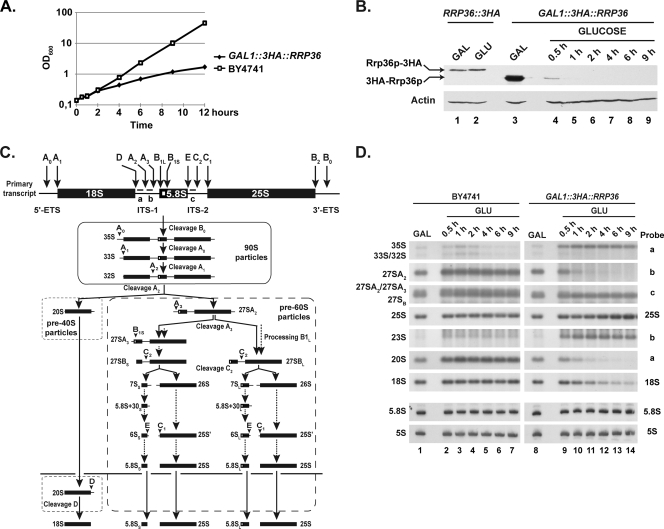
FIG. 3.
Rrp36p depletion affects early cleavages of the pre-rRNA and the production of the mature 18S rRNA in yeast cells. The GAL1::3HA::RRP36 or BY4741 (WT) strain was shifted from a galactose- to a glucose-based medium and maintained under conditions of exponential growth. (A) Growth curve of yeast cells undergoing Rrp36p depletion. Growth of the strains was followed by measuring the optical density of the cultures at 600 nm (OD600) at different times after the nutritional shift. (B) Depletion of 3HA-Rrp36p following the nutritional shift. Total proteins were extracted from the RRP36::3HA strain grown on galactose (GAL)- or glucose (GLU)-containing medium and from the GAL1::3HA::RRP36 strain before transfer to the glucose-based medium (GAL) and at different times after transfer to the glucose-based medium (GLUCOSE). The accumulation of 3HA-tagged Rrp36p proteins and actin (loading control) was assessed by Western blotting using anti-HA and actin-specific antibodies, respectively. Note that Rrp36p-3HA is slightly larger than 3HA-Rrp36p due to the insertion of a linker sequence, in addition to the 3HA tag, into the Rrp36p-3HA fusion protein. (C) Scheme of the pre-rRNA processing pathway in S. cerevisiae. Endonucleolytic cleavages and exonucleolytic trimmings are marked by solid and dotted arrows, respectively. The positions of the oligonucleotide probes used to detect the different pre-rRNAs analyzed in this study (a, b, and c) are shown on the initial precursor; their sequences are detailed in Table S1 in the supplemental material. (D) Early pre-rRNA processing defects in cells undergoing Rrp36p depletion. The BY4741 and GAL1::3HA::RRP36 strains were shifted from a galactose- to a glucose-based medium, and culture samples were collected before the nutritional shift (GAL) and at different times after the nutritional shift (GLU). Total RNAs were extracted from these cell samples, and the accumulation of the different (pre-)rRNAs was analyzed by Northern blotting using the indicated oligonucleotide probes (see Fig. 3C and Table S1 in the supplemental material).