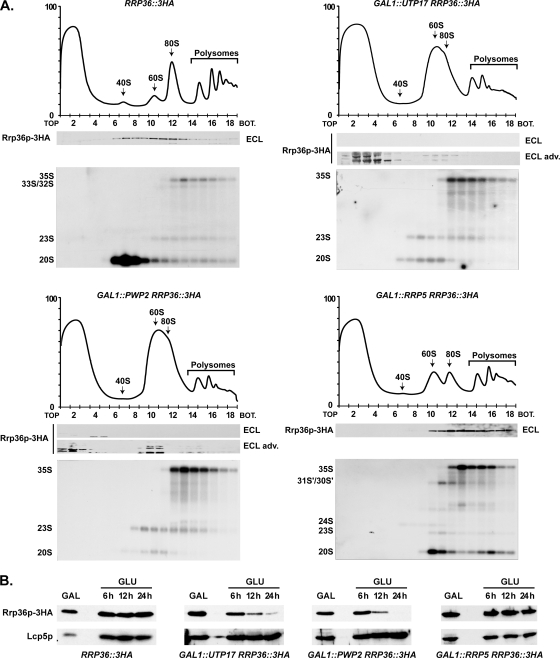
FIG. 5.
Components of the UTP-A and UTP-B complexes, but not Rrp5p, are required for the incorporation of Rrp36p into preribosomes. (A) Sedimentation profile of Rrp36p-3HA in WT cells (upper left panel) or in cells lacking Utp17p (upper right panel), Pwp2p (lower left panel), or Rrp5p (lower right panel). The GAL1::UTP17, GAL1::PWP2, and GAL1::RRP5 strains expressing Rrp36p-3HA were shifted from galactose- to glucose-containing medium and grown for 16, 18, or 14 h, respectively, to deplete the corresponding protein according to the method described in reference 25. As a control, the RRP36::3HA strain grown in the presence of glucose was used. Total extracts prepared from these cell samples were sedimented through sucrose gradients. The proteins contained in one-half of each fraction were analyzed by Western blotting using anti-HA antibodies to detect Rrp36p-3HA (note that Rrp36p-3HA was detected using regular ECL in WT and Rrp5p-depleted cells and the more sensitive “ECL Advance” [ECL adv.] kit in Utp17p- and Pwp2p-depleted cells). Total RNAs were extracted from the other half of each fraction, and the pre-rRNAs were detected by Northern blotting using probe a (Fig. 3C). BOT, bottom. (B) Accumulation levels of Rrp36p-3HA in WT or Utp17p-, Pwp2p-, or Rrp5p-depleted cells. The WT, GAL1::UTP17, GAL1::PWP2, and GAL1::RRP5 strains expressing Rrp36p-3HA were transferred from a galactose- to a glucose-containing medium, and cultures were maintained under conditions of exponential growth. Total protein extracts prepared from cell samples harvested before the nutritional shift (GAL) and 6, 12, and 24 h after the nutritional shift (GLU) were analyzed by Western blotting. Rrp36p-3HA was detected using anti-HA antibodies and regular ECL. Lcp5p was detected using specific antibodies.