Abstract
Development of broadly cross-reactive neutralizing antibodies (NAbs) remains a major goal of HIV-1 vaccine development, but most candidate envelope immunogens have had limited ability to cross-neutralize heterologous strains. To evaluate the immunogenicity of subtype A variants of HIV-1, rabbits were immunized with pairs of closely related subtype A envelopes from the same individual. In each immunogen pair, one variant was readily neutralized by a variety of monoclonal antibodies and plasma antibodies, while the other was neutralization resistant, suggesting differences in the exposures of key epitopes. The breadth of the antibody response was evaluated against subtype A, B, C, and D variants of HIV-1. The specificity of the immunogen-derived neutralizing antibody response was also compared to that of the infected individuals from whom these variants were cloned. None of the immunogens produced broad neutralizing antibodies in immunized animals, and most of the neutralizing antibodies were directed to the variable loops, particularly the V3 loop. No detectable antibodies to either of the potentially exposed conserved epitopes, the membrane proximal external region, or the CD4 binding site were found with immunized rabbits. In contrast, relatively little of the neutralizing activity within the plasma samples of the infected individuals was directed to linear epitopes within the variable loops. These data indicate that immunogens designed to expose conserved regions did not enhance generation of broadly neutralizing antibodies in comparison with the immunogens that failed to expose those regions using this immunization approach.
The ability to elicit broadly cross-reactive neutralizing antibodies (NAbs) is likely to be an important component of an effective vaccine to human immunodeficiency virus type 1 (HIV-1). Unfortunately, the HIV-1 envelope (Env)-based vaccines developed to date do not elicit such antibodies. Initial vaccines based on soluble, monomeric gp120 generated antibodies capable of only weakly neutralizing the homologous virus, with a very narrow breadth of cross-reactivity (13, 30, 53). Subsequent modifications to the Env immunogens, including variable loop deletions (15, 20, 31, 34, 35, 61, 64-66), alterations in the glycosylation pattern (4, 10, 11, 14, 30, 43, 55, 56), epitope repositioning (39, 46), the use of consensus Envs (22, 36, 37, 47), and the use of soluble trimeric gp140 molecules as immunogens (1-3, 5, 14, 16, 20, 21, 24, 25) have led to only modest enhancements in NAb breadth or potency. These modified Env immunogens have failed to redirect NAbs from the variable loops to more conserved regions of Env (reviewed in reference 33).
Differences in Env structure between HIV-1 subtypes may further hinder efforts to elicit broadly cross-reactive antibodies capable of protecting against transmitted strains worldwide. Most immunogens tested to date have been derived from subtype B Envs. However, there are clear antigenic differences between subtype B strains and the subtype A and C strains that account for most infections worldwide (6, 8, 27, 28, 40, 42). For instance, most transmitted subtype A Envs are resistant to the monoclonal antibodies 2G12, b12, 2F5, and 4E10, either because of alterations in the epitopes for these monoclonal antibodies (MAbs) or because the epitopes are shielded in these Envs (6, 8). It is therefore possible that even NAbs specific for a conserved region of subtype B Envs, such as the CD4 binding site, would not be able to access and neutralize a similar epitope on a subtype A Env.
In order to evaluate the immunogenicity of subtype A Envs, which account for ∼25% of global HIV-1 infections (12), we previously investigated the types of antibody responses elicited following gp160 priming and gp140 boosting with immunogens derived from four subtype A Envs in comparison to the subtype B Env SF162 (38). These experiments were also designed to explore whether deriving immunogens from HIV-1 Envs isolated from early in infection would better target NAbs to transmitted strains. Although all of the subtype A-based immunogens and the SF162 immunogen elicited anti-V3 NAbs capable of neutralizing the easy-to-neutralize SF162 pseudovirus, only one of the four immunogens generated homologous NAbs (38). Even immunogens with shorter variable loops or fewer potential N-linked glycosylation sites (PNGS) did not lead to enhanced breadth of neutralization against heterologous subtype A or B Envs (38). However, the four subtype A Envs used in these immunizations were generally neutralization resistant to both plasma samples from HIV-1-infected individuals and to monoclonal antibodies (6), raising the possibility that the poor breadth observed could be related to the shielding of conserved epitopes within these Envs.
In order to determine whether using subtype A Env immunogens that do not shield conserved epitopes could improve neutralization breadth, here we performed immunizations with pairs of Env immunogens derived from two individuals acutely infected with subtype A HIV-1. The Envs in each pair were very similar in their amino acid sequences yet differed dramatically in their neutralization phenotype (6, 9) (Fig. 1A). The pair from subject Q461 had a neutralization-resistant Env, Q461e2 (termed Q461e2R to indicate neutralization resistance), and a neutralization-sensitive Env, Q461d1 (termed Q461d1S to indicate neutralization sensitivity), which was sensitive to neutralization by plasma, 2F5, 4E10, b12, and soluble CD4 (sCD4). We previously demonstrated that the neutralization sensitivity of the Q461d1S Env is mediated entirely by two amino acid substitutions in gp41, one in the first heptad repeat and one in the membrane proximal external region (MPER) (9). These mutations led to enhanced exposure of both the CD4 binding site and the MPER (9). From subject Q168, the Env Q168b23S was sensitive to autologous and heterologous plasma and to the MPER antibodies 2F5 and 4E10 but resistant to b12 and sCD4, while Q168a2R was weakly neutralized by the MPER antibodies, less sensitive to neutralization by autologous plasma, and resistant to heterologous plasma (6). The Q168a2R and Q168b23S Envs contain identical sequences in the MPER region yet have >500-fold differences in neutralization sensitivity to 2F5 and 4E10, indicating that the exposure of the MPER region, rather than the sequence, likely accounts for the enhanced neutralization of the Q168b23S Env.
FIG. 1.
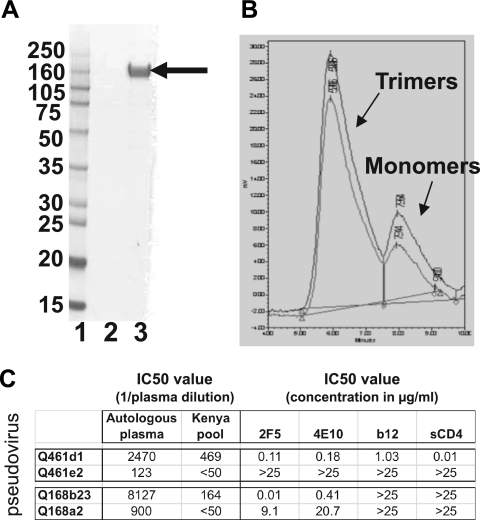
Analysis of Q461d1S gp140 used for immunizations. (A) SDS-PAGE analysis of final preparation of Q461d1S gp140 from the GNA capture and DEAE and CHAP columns. Lane 1 contains molecular weight standards, lane 2 the concentrated DEAE flowthrough, and lane 3 the final concentrated protein. The purified Q461d1S gp140 protein is indicated by an arrow. The sizes of the molecular weight markers (in thousands) are indicated on the left. (B) Binding of purified gp140 subtype A to CD4 as determined by a high-pressure liquid chromatography (HPLC)-based assay. The bottom line represents the protein obtained after the GNA column, and the top line represents purified protein after all three steps. The trimer and monomer peaks are marked. (C) Summary of neutralization characteristics of all four HIV-1 subtype A Env variants used in the immunizations, adapted from reference 6. The pseudovirus is shown in the far left column. IC50 values for plasma sample (left) and monoclonal antibodies (right) are displayed. The autologous plasma samples were taken 3.7 ypi for subject Q461 and 2.6 ypi for subject Q168. The Kenya pool was derived by pooling plasma from 30 HIV-1-infected individuals in Kenya and has been described previously (6).
Thus, to directly test whether using Env immunogens that expose conserved epitopes could enhance neutralization breadth immunization, here we immunized with these pairs of related Envs, in which one variant exposes conserved regions, while the other does not. We also compared the specificity of the NAb responses following immunization with these Envs with the specificities of the NAbs that developed during natural infection in the individuals from whom these variants were cloned.
MATERIALS AND METHODS
Subjects and envelope immunogens.
HIV-1 envelope variants and plasma samples were collected from two HIV-1 subtype A-infected subjects that were part of a prospective cohort study of high-risk women from Mombasa, Kenya, which was approved by the ethical review committees of the University of Nairobi, the University of Washington, and the Fred Hutchinson Cancer Research Center. The two pairs of HIV-1 envelopes used for this study were cloned directly from peripheral blood mononuclear cells (PBMCs) and have been described previously (48) (Fig. 1A). The pair from subject Q461 were cloned at an estimated 28 days postinfection (dpi), while the pair from subject Q168 were cloned from an estimated 23 dpi, based on HIV-1 RNA testing and serology as described previously (49-51).
Recombinant vaccinia viruses.
Recombinant vaccinia viruses encoding the full-length envelope open reading frame (encoding gp160 with natural cleavage sites intact) were constructed as previously described (32, 38). The full-length gp160 env gene was inserted into the thymidine kinase gene of vaccinia virus v-NY, which is a plaque-purified, replication-competent derivative of the New York City Board of Health strain of vaccinia virus. Expression of the env gene is under the control of a synthetic early-late promoter (17) and was verified by Western blot analysis (data not shown).
Construction of subtype A envelope expression plasmids.
The sequences encoding the open reading frame of the ectodomain of the Env protein from the HIV-1 subtype A isolates was codon modified as described elsewhere (29, 68) and constructed synthetically as a 2.1-kb EcoRI-XbaI DNA fragment. This gene cassette contained the protein-encoding region of the Env protein fused in frame to the human tissue plasminogen activator (tPA) signal sequence (18). In order to stabilize the oligomeric structure of the encoded gp140 protein, a series of mutations were introduced in the primary (REKR) and secondary protease (KAKRR) cleavage sites in the Env polypeptide (23). The resulting Env expression cassette (gp140) was cloned into the EcoRI-XbaI sites of the pCMV3 expression vector for transient transfection of 293 cells.
Expression and purification of soluble gp140.
The gp140 versions of Envs used as immunogens were expressed transiently by bulk transfecting HEK293 cells at 4L scale. The media were collected 72 to 96 h after transfection, concentrated 10 times, filtered using a 0.22-μM filter, centrifuged, and processed for purification. A three-step strategy was designed for efficient purification of soluble oligomeric gp140 trimers from subtype A variants. First, concentrated supernatants obtained from transiently transfected HEK293 cells were passed over a Galanthus nivalis agarose (GNA) lectin column. The oligomeric gp140 protein was bound to the column, and most contaminating proteins flowed through. The bound oligomeric gp140 was eluted with 500 mM methyl mannose pyranoside (MMP). Next, the captured oligomeric gp140 protein was passed over DEAE and ceramic hydroxyapatite (CHAP) columns, allowing oligomeric gp140 to flow through both columns and contaminating proteins to bind to one column or the other. All the column fractions were analyzed on SDS-PAGE and also in a CD4 receptor-binding assay as described elsewhere in detail (58-60).
Immunization.
Groups of six specific-pathogen-free New Zealand White rabbits (Oryctolagus cuniculus) of either sex, 5 to 7 months old, were immunized at 0 and 8 weeks by skin scarification with 5 × 107 PFU of one of the four recombinant vaccinia viruses expressing the full-length gp160 and processed forms of the Env. Rabbits were boosted at week 24 with 100 μg of the corresponding Env gp140 produced in HEK cells as described above, formulated in incomplete Freund's adjuvant, and delivered intramuscularly. Serum samples were collected both prior to (prebleeds) and 2 weeks following immunization to assess neutralization activity.
Peptides.
Peptides derived from the variable regions of the Q461d1S and Q461e2R Envs were used for epitope mapping experiments. These Envs were identical in their variable loop sequences, except for one amino acid in the V2 C crown; that peptide sequence matched that of the Q461e2R Env. This amino acid difference did not contribute to changes in the neutralization sensitivity between the Envs (9). The peptides were CTDWTNNATSTNQTTPATSE (V1 N terminus, derived from the amino-terminal side of the V1 loops), PATSEETGVKNCSFNITTEL (V1V2 junction [jxn], derived from the junction between the V1 and V2 loops), ITTELRDKKQKVYSLFYKLD (V2 N crown, derived from the amino terminus of the central region or “crown” of the V2 loop), FYKLDVVQISESNSSNSSNF (V2 C crown, derived from the carboxy-terminal side of the V2 crown), NSSNFTQYRLINCNTSAITQ (derived from the C terminus of the V2 loop), CIRPGNNTRKSVRIGPGQ (V3 N terminus, derived from the amino terminus of the V3 loop), and PGQAFYATGDITGDIRNAHC (V3 C terminus, derived from the carboxy terminus of the V3 loop). Peptides to the canonical 2F5 (NEQELLELDKWASLWN) and 4E10 (NWFDITNWLWYIRKKK) epitopes were also used.
Binding assays.
Binding to purified trimeric gp140 protein derived from four HIV-1 subtype A Envs was assessed in a Luminex-based platform. Envs used for Luminex experiments were produced using 293 suspension cells and purified as described previously (57), with one additional gel filtration step with Superdex 200PG (GE Healthcare) to resolve the trimers. Purified, trimeric clade A envelopes (12 μg/reaction mixture[rxn]) were coupled to Bioplex beads according to the manufacturer's instructions. Beads coupled to Q168a2R, Q259d2.17, Q461e2R, and Q769h5 gp140 trimers were mixed at 750 beads/μl for each protein. Using a multiscreen BV 1.2-μm plate (Millipore), the beads were washed 2 times with 1× PBS, mixed with serially diluted rabbit serum, and incubated 60 min at room temperature (RT), with shaking at 600 rpm. Serum was aspirated with suction, and the beads were washed 3 times with 1× PBS and 0.1% Tween 20. Goat anti-rabbit RPE (Southern Biotech), 1:500, was added and incubated as mentioned above. The beads were washed 3 times with PBS and Tween 20 and then 1 time with PBS and run on the Bioplex 200 (Bio-Rad). Endpoint titers were defined as the lowest dilution of plasma in which binding was threefold greater than the highest binding observed with the prebleed serum samples.
Neutralization assays.
Neutralization was assessed in triplicate in at least 2 independent experiments using the TZM-bl neutralization assay as described previously (6, 54, 63). Briefly, pseudotyped viral particles were generated by transfecting plasmid DNA containing a full-length gp160 env gene into 293T cells along with an envelope-deficient HIV-1 subtype A proviral plasmid, Q23Δenv, as described previously (48). Then, 500 infectious particles of pseudovirus, as determined by infection of TZM-bl cells, were incubated with serial dilutions of plasma, sera, or monoclonal antibody for 1 h. TZM-bl indicator cells were added, and infection levels were determined by assessing β-galactosidase activity after 48 h. Median inhibitory concentrations (IC50) were defined as the reciprocal dilution of plasma/sera or concentration of monoclonal antibody that resulted in 50% inhibition, calculated, as described previously, using the linear portion of the neutralization curve (6, 63). The “Kenya plasma pool” collected from 30 HIV-1-infected individuals in Kenya between 1998 and 2000 has been described previously (6).
Peptide competition during neutralization.
Serial dilutions of 20 μl of postimmune serum samples or subject plasma were incubated in triplicate with an equal volume of peptide at 10 μg/ml for 1 h at 37°C, and 20 μl containing 500 infectious particles of pseudovirus was added and incubated for an additional hour at 37°C. The wells containing pseudovirus alone or cells alone in the absence of sera or plasma were also incubated with peptide in order to control for the direct effects of the peptide on viral entry. The serum/peptide/pseudovirus mixture was incubated with DEAE-dextran-treated TZM-bl cells exactly as performed for a standard neutralization assay. The percentage of reduction in neutralization was determined by calculating the area under the curve (AUC) in the presence or absence of peptide. The percentage of contribution of a given peptide to neutralizing activity was then calculated as 100·(AUC in the absence of peptide − AUC with peptide)/(AUC in the absence of peptide).
Detection of antibodies to the CD4 binding site.
Detection of CD4 binding-site antibodies was performed as described previously (57). Briefly, rabbit serum samples or HIV-1-infected subject plasma at a 1:10 dilution were incubated for 3 h with gentle rotation with tosylactivated MyOne Dynabeads (Invitrogen) coated with either wild-type SF162 gp120 protein or SF162 gp120 protein containing a D368R mutation which abolishes binding to CD4 (gp120bs). Depletions were repeated three times in order to completely remove the anti-gp120 antibodies. The beads were then removed with a magnet. Antibodies were eluted from the beads in a series of increasingly acidic solutions as previously described (44, 57). The antibody titers of depleted sera/plasma or eluted antibodies were assessed by enzyme-linked immunosorbent assay (ELISA) using soluble SF162 gp120 and soluble SF162 D368R gp120 (gp120bs) as previously described (57). Neutralization assays with the depleted plasma and eluted antibodies were performed using the TZM-bl neutralization assay as described above.
RESULTS
Immunization.
Groups of six animals per immunogen were primed at 0 and 8 weeks by skin scarification with 5 × 107 PFU recombinant vaccinia viruses expressing the full-length gp160 and cleaved forms of the Q461d1S, Q461e2R, Q168b23S, or Q168a2R Env. The animals received a boost at week 24 with 100 μg of the corresponding Env gp140. Prior to boosting, the purity of the gp140 products was determined by SDS-PAGE, Western blotting, size exclusion column chromatography (SEC), and a CD4 binding assay, as shown in Fig. 1 for the Q461d1S Env as a representative example. The purified protein was >95% pure as determined by SDS-PAGE (Fig. 1A), and >85% of the protein was estimated to be in its trimeric conformation (Fig. 1B). The neutralization properties of all of the Envs used are summarized in Fig. 1C.
Development of antibodies capable of binding to HIV-1.
Two weeks after booster immunization with the recombinant gp140 Envs, the serum samples from each immunization group were pooled and tested for their ability to bind to trimeric gp140 proteins from four different HIV-1 Envs. Each immunogen resulted in the development of antibodies capable of binding to the Q461e2R and Q168a2R gp140 trimers, and the titers were similar whether the rabbits received a homologous or heterologous immunogen (Table 1). Antibodies capable of binding to the heterologous Q259d2.17 and Q769h5 gp140 trimers were also present in all immunization groups, but titers were 5-fold to 25-fold lower against these proteins (Table 1). Overall, there were no significant differences between immunization groups in the ability to bind to any of the gp140 proteins by Wilcoxon's signed-rank test.
TABLE 1.
Endpoint titers of binding antibodies to four different HIV-1 gp140 proteins
| Protein tested | Endpoint titer (1/serum dilution)a |
|||
|---|---|---|---|---|
| Q461d1S immunized | Q461e2R immunized | Q168b23S immunized | Q168a2R immunized | |
| Q461e2R gp140 | 62,500 | 62,500 | 62,500 | 12,500 |
| Q168a2R gp140 | 62,500 | 62,500 | 312,500 | 62,500 |
| Q259d2.17 gp140 | 12,500 | 12,500 | 62,500 | 12,500 |
| Q769h5 gp140 | 12,500 | 2,500 | 12,500 | 2,500 |
Numbers in boldface type indicate homologous binding titers.
Development of homologous and heterologous NAbs to HIV-1.
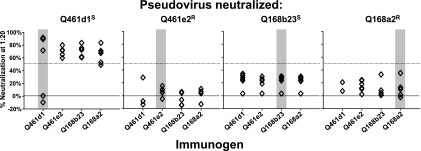
Serum samples from the immunized rabbits were tested for their ability to neutralize pseudoviruses expressing homologous and heterologous Env variants (Fig. 2). Only four of the six animals immunized with the Q461d1S immunogen mounted homologous NAb responses to the neutralization-sensitive Q461d1S pseudoviruses (Fig. 2, first panel, gray bar). In order to ensure that the two animals lacking neutralizing antibodies had received appropriate immunization, the HIV-1 Env binding activity of serum samples from the individual animals within this group was assessed (see Table S1 in the supplemental material). All six of the animals generated HIV-1-specific antibodies following immunization, though the two animals lacking neutralizing activity against the Q461d1S pseudovirus, WA549 and WA550, had ∼100-fold-lower binding titers to gp140 trimers (see Table S1). Therefore, we could not rule out a problem in the immunization of these two animals.
FIG. 2.
Homologous and heterologous neutralizing antibody responses against pseudotyped viruses made with the four env variants used as the basis for immunogen design. The ability of a 1:20 dilution of rabbit serum samples to neutralize the pseudoviruses shown at the top following DNA priming and gp140 boosting is indicated along the y axis. Each point represents the immune response by an individual rabbit, with six rabbits per immunogen. The immunogen is indicated along the bottom x axis. Homologous responses are highlighted by the gray bars. Fifty percent neutralization is indicated by the dotted horizontal line.
All of the animals in the Q461e2R, Q168b23S, and Q168a2R imunogen groups could neutralize the Q461d1S pseudovirus at 50% or greater, consistent with the extreme neutralization sensitivity of this Env variant (6, 9) (Fig. 2). There were no significant differences between immunogen groups in their ability to neutralize the Q461d1S pseudovirus as determined by an unpaired t test.
None of the animals in the Q461e2R, Q168b23S, and Q168a2R immunogen groups could neutralize their homologous Env pseudoviruses (Fig. 2). In each case, <50% neutralization was observed at a 1:20 serum dilution; use of higher serum dilutions led to nonspecific neutralization that did not differ from levels found in prebleed serum samples (data not shown). We also did not see evidence of heterologous neutralization, aside from the neutralization of Q461d1S pseudovirus. None of the animals in any immunization group could neutralize the Q461e2R, Q168b23S, or Q168a2R pseudovirus (Fig. 2). To more thoroughly assess the ability of serum samples from these rabbits to assess heterologous strains of HIV-1, a serum sample from each animal was tested for its ability to neutralize an expanded panel of 14 pseudoviruses representing Envs from subtypes A, A/D, B, C, and D (6-8, 41, 42). These pseudoviruses have a range of neutralization sensitivities, as demonstrated by the range in IC50 values for a pool of plasma samples from HIV-1-infected individuals in Kenya (Table 2), and would represent both tier 1 and tier 2 viruses (52). None of the animals in any immunization group were able to neutralize this expanded panel of pseudoviruses >50% with a 1:20 dilution of serum; higher concentrations of serum samples resulted in only nonspecific neutralization that did not differ from levels found in prebleed serum samples (Table 2). Thus, no breadth of neutralization was observed following immunization with any of these envelope-based immunogens.
TABLE 2.
Breadth of neutralizing antibody responses of immunized rabbits
| Pseudovirus testeda | Subtype | IC50b |
||||
|---|---|---|---|---|---|---|
| Kenya poolc | Q461d1S immunizedd | Q461e2R immunizedd | Q168b23S immunizedd | Q168a2R immunizedd | ||
| Q769b9 | A | <50 | <20 | <20 | <20 | <20 |
| QF495.23 M.ENV.A1 | A | 100 | <20 | <20 | <20 | <20 |
| Q842d16 | A | 135 | <20 | <20 | <20 | <20 |
| BJ613.E1 | A | 142 | <20 | <20 | <20 | <20 |
| BS208.B1 | A | 207 | <20 | <20 | <20 | <20 |
| BF535.A1 | A/D | 76 | <20 | <20 | <20 | <20 |
| QA790.204I.ENV.C1 | A/D | 148 | <20 | <20 | <20 | <20 |
| THRO4156.18 | B | 52 | <20 | <20 | <20 | <20 |
| 6434.4 | B | 106 | <20 | <20 | <20 | <20 |
| QB099.227 M.ENV.C8 | C | 108 | <20 | <20 | <20 | <20 |
| QC406.70 M.ENV.F3 | C | 119 | <20 | <20 | <20 | <20 |
| Du156.12 | C | 137 | <20 | <20 | <20 | <20 |
| QA465.59 M.ENV.D1 | D | <50 | <20 | <20 | <20 | <20 |
| QD435.100 M.ENV.A4 | D | 69 | <20 | <20 | <20 | <20 |
The pseudoviruses used to assess neutralization breadth have been described previously (6, 41, 42).
The IC50 value is defined as the reciprocal dilution of sera which mediates 50% neutralization; <20 indicates that <50% neutralization was observed with a 1:20 dilution of plasma.
The Kenya pool was derived by pooling plasma from 30 HIV-1-infected individuals in Kenya as previously described (6).
Each of six animals in each immunization group was tested individually.
Development of NAbs to the V1/V2 and V3 loops in immunized rabbits.
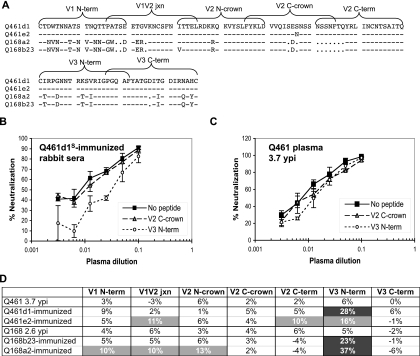
To determine whether the neutralization of the Q461d1S pseudovirus by immunized rabbits was accounted for by NAbs specific to the variable loops, peptide competition neutralization assays were performed (Fig. 3). The specificities of the NAbs following immunization were also compared to the specificities of the NAbs formed during natural infection in the individuals from whom these env variants were cloned. These assays used 20-mer peptides to the V1/V2 and V3 loops of the Q461d1S Env, except for the V2 C-crown peptide, which matched the sequence of the Q461e2R Env (Fig. 3A). The serine-to-asparagine change in the V2 C crown did not contribute to the changes in neutralization phenotype between these variants (9; data not shown). A reduction in neutralization when serum or plasma samples was preincubated with these peptides was used to determine the contribution of antibodies to that peptide region to the overall neutralizing potential of a serum sample. Examples of the curves used to calculate the contribution of NAbs to a specific region are shown in Fig. 3B and C.
FIG. 3.
Contribution of antibodies to the V1, V2, and V3 loops to the neutralizing potential of plasma from HIV-1-infected subjects and serum samples from rabbits immunized against the Q461d1S pseudovirus. (A) Amino acid alignment of the V1V2 loops (top) and V3 loops (bottom) for the four Env variants used as immunogens is shown. The location of the 20-mer peptides used for peptide competition neutralization assays are indicated by the brackets above the sequence; the sequences were made to match the Q461e2R variant, which is identical to the Q461d1S sequence, except of a single amino acid within the V2 C crown. Dashes indicate that the same amino acid was present at that position, and dots indicate a deletion of an amino acid at that position. (B) Example of representative neutralization curves depicting a decrease in neutralization by preincubation of serum samples from Q461d1S-immunized rabbits with V3 N-terminal, but not V2 C-crown, peptide. (C) Example of representative neutralization curves depicting minimal reduction in neutralization by preincubation of plasma from subject Q461 at 3.7 ypi with V2 C-crown and V3 N-terminal peptides. (D) The plasma or serum sample indicated in the far left column were preincubated with the peptide indicated along the top before the addition of the Q461d1 pseudovirus in a TZM-bl neutralization assay. The percentage of contribution of the indicated peptide to the neutralizing activity of that serum or plasma sample is indicated in the table. Increasing contribution to neutralizing activity is indicated by darker shaded boxes. Unshaded boxes indicate <5% contribution of that epitope to neutralizing activity.
NAbs specific to the amino terminus of the V3 loop accounted for a large proportion of the neutralizing activity of the immunized rabbit serum samples against the Q461d1S pseudovirus, ranging from 16% in the Q461e2R-immunized animals to 37% in the Q168a2R-immunized animals (Fig. 3D). Smaller proportions of NAbs to specific targets within the V1/V2 loops contributed to neutralization of the Q461d1S pseudovirus in these animals, with generally <10% of the antibodies in Q461d1S-, Q461e2R-, and Q168b23S-immunized animals specific for any of the five regions of the V1 and V2 loops tested (Fig. 3D). The Q168a2R-immunized animals had slightly more V1/V2 antibodies, with 10% of the antibodies directed to the V1 N terminus, 10% to the V1V2 junction, and 13% to the V2 N crown (Fig. 3D). Despite the sequence differences between the variable loops (Fig. 3A), small proportions of antibodies specific for the V1/V2 and V3 loops of the Q461d1S pseudovirus were generated following immunization with Q168a2R and Q168b23S immunogens. This is consistent with recent data indicating that antibodies to variable loops can still cross-neutralize heterologous strains (62). Overall, approximately half of the neutralizing activity of these serum samples appeared to be directed to linear epitopes within the V1/V2 and V3 loops, and no significant differences in the contributions of the variable loop antibodies were observed following immunization with any of the immunogens.
Development of NAbs to the V1/V2 and V3 loops in infected subjects.
To examine how the antibody response in immunized rabbits compared to the antibodies elicited in the infected individuals from whom these env variants were cloned, the specificities of the NAbs in the plasma samples of the infected subjects were evaluated. Both subjects from whom these env variants were cloned had plasma antibodies capable of neutralizing heterologous strains of HIV-1 (6). However, few of the NAbs during natural infection appeared to be directed to the variable loops; examples of the curves for the immunized rabbits in comparison to those for the infected individuals are shown in Fig. 3B and C. Neither infected subject developed a >10% fraction of NAbs specific for any of the linear peptide epitopes explored (Fig. 3D). Overall, less than one quarter of the neutralizing activity of these plasma samples against the Q461d1S Env was accounted for by NAbs to linear epitopes in the V1/V2 and V3 loops.
Development of NAbs to the MPER region.
In order to explore whether some of the unaccounted neutralizing activity of the rabbit serum samples was directed to the MPER region, particularly with the Q461d1S- and Q168b23S-immunized rabbits, the titers of antibodies specific for the canonical 2F5 and 4E10 epitopes were assessed. Endpoint titers were >20 for all immunization groups, indicating no significant detection of MPER-specific antibodies. These results were confirmed in peptide competition neutralization assays using peptides matching the 2F5 and 4E10 epitopes. Although these peptides could be used to specifically block neutralization by 2F5 or 4E10, but not by other antibodies, such as b12, none of the immunogens generated antibodies to either the 2F5 or 4E10 epitope (not shown). Furthermore, 2F5 or 4E10-like NAbs were not detected with the plasma samples from either subject Q461 or Q168 (not shown).
Development of NAbs to the CD4 binding site in subject Q461.
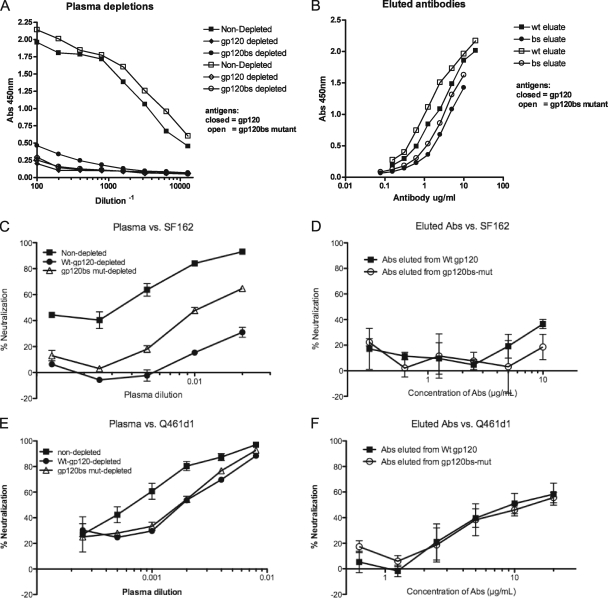
The Q461d1S Env is unusual for subtype A HIV-1 Env, in that it exposes the CD4 binding site to antibodies such as b12, unlike the Q461e2R Env and most other subtype A Envs (9). In addition, plasma antibodies from this subject were capable of heterologous neutralization, so we evaluated the contribution of antibodies specific for the CD4 binding site in the plasma sample from subject Q461 by performing absorption experiments using wild-type SF162 gp120 and mutant SF162 gp120 that prevent the binding of CD4bs-specific antibodies (gp120bs), as described previously (44, 57). Depletion of the 3.7-year postinfection (ypi) Q461 plasma sample with the wild-type SF162 gp120 protein coupled to magnetic beads effectively removed most gp120-binding antibodies, while a small fraction remained in the plasma following depletion with beads coated with the gp120bs mutant (Fig. 4A). These residual antibodies could not bind the mutant gp120bs protein (Fig. 4A, open symbols). Thus, a small fraction of the gp120-specific antibodies in this subject's plasma were specific for the CD4 binding site. To confirm this further, the antibodies bound to the beads were eluted and tested for their ability to bind to gp120 proteins. Fewer gp120-binding antibodies were eluted from the mutant gp120bs-coated beads than from the wild-type gp120-coated beads, consistent with the presence of CD4 binding site-specific antibodies in this plasma sample (Fig. 4B). These results were completely reproducible in a second, independent experiment (data not shown).
FIG. 4.
Contribution of antibodies to the CD4 binding site plasma from subject Q461. (A) Plasma from subject Q461 at 3.7 ypi was preincubated with magnetic beads coated with bovine serum albumin (BSA) (squares), wild-type SF162 gp120 (diamonds), or SF162gp120 containing mutations in the CD4 binding site (circles). The beads were removed, and the depleted plasma samples were tested for their ability to bind to wild-type SF162 gp120 (closed symbols) or CD4 binding site mutant gp120 (open symbols). The absorbance at 450 nm is shown on the y axis, with the serum dilution on the x axis. (B) Antibodies eluted from the wild-type SF162 gp120 beads (squares) or the CD4 binding site mutant (circles) were tested for their ability to bind to wild-type gp120 protein (closed) or gp120bs mutant protein (open). The absorbance at 450 nm is shown on the y axis, with the concentration of antibody on the y axis. (C) Plasma (represented in panel A) that was nondepleted (squares), depleted with wild-type SF162 gp120 protein (closed circles), or depleted with beads coated with CD4 binding site mutant gp120 protein (open triangles) were tested for their ability to neutralize SF162 pseudovirus. The percentage of neutralization is shown on the x axis, with the plasma dilution on the y axis. (D) The antibodies eluted from the Wt-gp120 or BS-mut gp120 beads, represented in panel B, were tested for their ability to neutralize the SF162 pseudovirus. (E) Ability of depleted plasma to neutralize Q461d1S pseudovirus, described in the legend for panel C. (F) Ability of antibodies eluted from Wt- or BS-mut-coated beads to neutralize Q461d1S pseudovirus, as described in the legend for panel D. All results are representative of two independent experiments; contribution of neutralization was calculated according to the AUC, as described in Materials and Methods.
In order to determine whether the CD4 binding site-specific antibodies in subject Q461's plasma could mediate neutralization, the ability of the depleted plasmas and eluted antibodies to neutralize HIV-1 pseudoviruses was tested. Depletion of the Q461 plasma with wild-type gp120-coated beads removed ∼86% of the neutralizing activity of the plasma against the SF162 pseudovirus (Fig. 4C), though the eluted antibodies were of such low concentration they did not demonstrate clear neutralization of the SF162 pseudovirus (Fig. 4D). However, depletion with the gp120bs beads reduced neutralization by only ∼59% (Fig. 4C), suggesting that CD4 binding site-specific antibodies account for a portion of the neutralizing antibodies to the heterologous SF162 pseudovirus.
In order to determine whether CD4 binding site-specific antibodies contributed to the neutralization of other viruses, including the autologous Q461d1S pseudovirus, the ability of the depleted plasma samples to neutralize Q461d1S and Q842d16 pseudoviruses was assessed. Only ∼25% of the neutralizing activity of the Q461 plasma was absorbed by depletion with wild-type SF162 gp120-coated beads, indicating that gp120-specific antibodies within this plasma sample were either low in frequency or not directed to epitopes shared by the SF162 gp120 protein (Fig. 4E). Similarly, only ∼25% of the activity was absorbed by the SF162 gp120bs mutant, indicating that CD4 binding site-specific antibodies did not account for the neutralization of the autologous Q461d1S pseudovirus (Fig. 4E). In addition, no difference in the neutralization of the Q461d1S pseudovirus was observed between antibodies eluted from the wild-type or gp120bs-coated beads (Fig. 4F). Similarly, no CD4 binding site-specific antibodies contributed to the ability of the Q461 plasma to neutralize the heterologous subtype A pseudovirus Q842d16 (data not shown). Thus, although antibodies specific to the CD4 binding site may account for a fraction of the neutralization of the SF162 pseudovirus by the Q461 plasma, CD4 binding site-specific antibodies did not appear to account for the neutralization of the autologous or one heterologous subtype A HIV-1 variant.
Development of NAbs to the CD4 binding site in rabbits immunized with Q461d1S and Q461e2R.
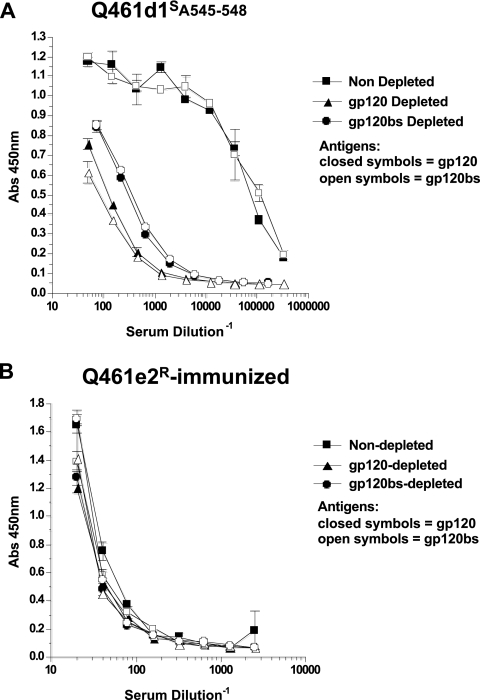
To determine whether immunization with either of the two immunogens derived from early env variants in subject Q461 could lead to antibodies specific for the CD4 binding site, the serum samples from the Q461d1S- and Q461e2R-immunized animals were similarly tested for the presence of antibodies to the CD4 binding site by absorption with wild-type gp120 or a gp120bs mutant. As found in the infected subject's plasma, in the Q461d1S-immunized animals, absorption with wild-type SF162 gp120 removed a large proportion of the binding antibodies, indicating that many of the antibodies in these immunized animals were directed to gp120 (Fig. 5A, squares versus triangles). Following absorption with the gp120bs-coated beads, some residual binding activity remained in the rabbit serum samples (Fig. 5A, circles versus triangles,). However, the residual antibodies, following gp120bs-coated bead absorption, bound similarly to both the gp120 wild-type and mutant gp120bs proteins, indicating that these antibodies were not specific to the CD4 binding site (Fig. 5A, open versus closed circles). This was distinct from the activity noted with the plasma from the infected subject, in which the residual antibodies following absorption with the gp120bs-coated beads could bind to wild-type (closed circles) but not mutant (open circles) gp120 protein (Fig. 4A). Thus, the Q461d1S-immunized rabbits did not show evidence of CD4 binding site-specific antibodies. Similarly, the Q461e2R-immunized animals did not show evidence of CD4 binding site-specific antibodies, though titers of antibodies to SF162 gp120 were much lower than those in Q461d1S-immunized animals (Fig. 5B).
FIG. 5.
Contribution of binding antibodies to the CD4 binding site within serum samples of rabbits immunized with the Q461d1S or Q461e2R Envs. Rabbit serum samples from Q461d1S-immunized animals (A) or Q461e2R-immunized animals (B) at a 1:10 dilution were preincubated with magnetic beads coated with BSA (squares), wild-type SF162 gp120 (triangles), or SF162gp120 containing mutations in the CD4 binding site (circles). The beads were removed, and the depleted plasmas were tested for their ability to bind to wild-type SF162 gp120 (closed symbols) or CD4 binding site mutant gp120 (open symbols). The absorbance at 450 nm is shown on the y axis, with the serum dilution on the x axis.
DISCUSSION
We performed immunizations with two pairs of related subtype A HIV-1 Envs, with very different neutralization phenotypes to directly address (i) whether HIV-1 Env variants that expose conserved neutralization epitopes can lead to enhanced formation of NAbs to those epitopes and (ii) the immunogenicity and specificity of Env immunogens from acute subtype A infection in comparison with the NAbs formed in the plasma of the individuals infected with those variants. Most of the NAbs in the immunized animals were directed to linear epitopes in the variable loops, particularly the V3 loop, and none of the immunization strategies produced broad NAbs. Furthermore, no detectable antibodies to the MPER region or to the CD4 binding site were found with immunized animals. Thus, even immunogens derived from Envs that are highly neutralization sensitive by virtue of overall conformational changes to expose existing epitopes (9) are not necessarily able to elicit antibodies to those epitopes, at least in the immunization protocol used here. In contrast, the individuals from whom these Env variants were cloned developed NAbs capable of neutralizing heterologous strains with specificities to epitopes that were primarily outside the variable loops or to conformational epitopes within the variable loops.
These immunogens were selected primarily to test whether early variants that potentially expose conserved neutralization epitopes would better direct NAbs to conserved regions of transmitted variants, particularly subtype A variants. Neither of the two Envs tested from each subject led to enhanced NAb breadth; in fact, three of the four immunogens tested did not even generate homologous NAb responses, despite eliciting high titers of HIV-1-specific binding antibodies. Furthermore, only 4/6 of the Q461d1S-immunized animals generated homologous NAbs to the Q461d1S pseudovirus, which is generally easy to neutralize. The two animals without neutralizing activity against the Q461d1S pseudovirus were the only animals with low binding antibody titers to HIV-1, suggesting that immunization in these animals was not robust.
Overall, none of the immunizations approximated the ability of NAbs from the infected subjects to neutralize heterologous viruses at 2 to 4 ypi (6). Unlike immunization, natural infection results in very high antigen burdens and continued diversification of that antigen pool, both of which could have contributed to the development of NAbs capable of heterologous neutralization in subjects Q461 and Q168. As intervening samples were not available from these subjects, it was not possible to determine when these subjects developed NAbs capable of heterologous neutralization and which Env variants elicited that NAb response. Thus, the failure of this immunization strategy to generate heterologous NAbs may reflect the fact that the early Env variants studied did not contribute to the generation of heterologous NAbs or that this immunization strategy was inadequate to mimic the responses that developed over time with high antigen loads in these subjects, or both.
It remains possible that these subtype A Env immunogens would have performed better if the immunization strategy were altered, for instance, by using different gp160 or gp140 immunogens, different adjuvants, or a virus-like particle approach. Other adjuvant/gp140 immunogen combinations used have been associated with better NAb breadth than what we observed in this study, though generally only at a 1:5 dilution of serum (45, 67), a concentration with which we observed only nonspecific inhibition of viral entry. Interestingly, one of the Env immunogens that led to broad NAb responses in a prior study was a neutralization-sensitive variant derived from an individual with broadly neutralizing plasma, though this Env was unusual in that it was capable of CD4-independent entry (67). Recognition of these epitopes in rabbits may have been different than it would have been with primate species expressing primate CD4 molecules (26). Overall, it is not clear whether the gp160 or gp140 immunogens themselves, the adjuvants, the delivery vectors, the species immunized, or even the methods used to assess breadth accounted for the poor breadth observed here compared to that for some other gp140 immunogens.
Following immunization with any of the Env immunogens, approximately half of the neutralizing activity against the Q461d1S pseudovirus could be competed with peptides to the variable loops. Since the Q461e2R and Q461d1S Envs are nearly identical in variable loop sequences, this indicates that the majority of the homologous neutralizing activity was directed against linear epitopes in the variable loops, particularly V3, as we have previously observed (19, 20, 38). In addition, despite significant differences in the variable loop sequences between the Q168a2R and Q168b23S immunogens and the Q461d1S pseudovirus, cross-reactive antibodies to the variable loops of the Q461d1S pseudovirus formed following Q168a2R and Q168b23S immunization. Most of these antibodies were specific for the amino terminus of the V3 loop, in which several regions of homology remain, including the GPGQ crown. Similar V3-specific cross-reactive antibodies following immunization have been reported previously (45).
Some of the neutralizing activity of the rabbit serum samples following immunization could not be accounted for by linear V1/V2 or V3 epitopes, indicating that the remainder of the neutralizing activity was likely directed to other regions of the Env or to conformational epitopes. The remaining neutralizing activity did not appear to be directed against either the 2F5 or 4E10 region of the MPER or the CD4 binding site, even for the animals in which the MPER (Q461d1S and Q168b23S immunized) or the CD4 binding site (Q461d1S immunized) was potentially exposed in the immunogen. Thus, other regions of the Env appear to be immunodominant.
In contrast to the NAbs observed following immunization, the majority of the plasma NAbs from the infected subjects from whom the Envs were cloned were directed against epitopes other than linear spans of the variable loops. We could not detect MPER-specific 2F5- or 4E10-like NAbs with these subjects. However, a proportion of the heterologous neutralization of the SF162 pseudovirus by subject Q461 was accounted for by NAbs specific to the CD4 binding site. Such CD4 binding site-specific NAbs significantly contribute to the broad heterologous neutralizing activity observed with a subset of subtype B-infected individuals (44, 57). However, in this subtype A-infected subject, CD4 binding site-specific antibodies did not necessarily account for much of the heterologous neutralizing activity, as neutralization of two other pseudoviruses, Q461d1S and Q842d16, was not mediated by CD4 binding site-specific antibodies. It remains unclear whether the heterologous neutralization observed with these plasma samples from 2 to 4 ypi was caused by multiple antibodies to multiple different epitopes or by one or a few antibodies with cross-reactive specificity. The recent identification of broadly neutralizing MAbs specific for conserved regions within the variable loops from an subtype A-infected individual highlight the facts that such broad NAbs need not be directed to the CD4 binding site or to the MPER and that broad neutralizing activity appears to frequently arise from a few antibodies with broad specificity (62).
One major caveat to these studies is that we still do not know how closely the structures of the Env proteins expressed by recombinant vaccinia viruses or the soluble gp140 immunogens resembled those of the pseudoviral particles whose neutralization phenotype was originally characterized. Further structural characterization of these immunogens has been hindered by the fact that reliable methods to compare antibody bindings between intact, membrane-bound gp160 molecules and gp140 molecules are not readily available. Furthermore, antibody binding to membrane-bound gp160 does not always correlate with neutralization susceptibility, suggesting that binding studies may not be a reliable surrogate marker for the exposure of particular neutralization epitopes on the immunogens (E. Lovelace and J. Overbaugh, unpublished data). Indeed, in this study, the immunogen structure may be particularly relevant to evaluating the effect of the Q461d1S gp140 immunogen because the neutralization susceptibility of this variant is determined by amino acid changes in gp41, including a threonine-to-alanine change in the first heptad repeat and an isoleucine-to-valine change in the MPER-induced conformation, changes leading to the exposure of the MPER and the CD4 binding site within this Env (9), and the gp140 was truncated proximal to the isoleucine-to-valine change. This truncation could have mitigated the effect of these key gp41 amino acid determinants, and it therefore remains possible that a Q461d1S immunogen, such as a virus like-particle, would have performed differently. This concern regarding gp140 immunogens, while of particular relevance here, is potentially a more general issue, and future studies that better define the antigenic properties of gp140 versus gp120/gp41 trimers in the viral context are needed to address this.
Overall, these data highlight challenges in devising a vaccine capable of generating potent, broadly neutralizing activity to the diverse strains and subtypes of HIV-1. Using subtype A Env immunogens, we were unable to approximate the potency or breadth of the NAb response in the subjects from whom the Envs were cloned using a gp160 prime and gp140 boost. Following immunization, but not natural infection, most NAbs were directed to the immunodominant V3 loop regardless of the envelope immunogen used. Thus, the results of the current study indicate that immunogens that exposed conserved regions, such as the CD4 binding site or the MPER, did not enhance generation of broadly neutralizing antibodies in comparison with the immunogens that failed to expose those regions using this immunization approach.
Supplementary Material
Acknowledgments
We thank Nancy Haigwood for helpful input on the study design, For Yue Tso for assistance in the construction and characterization of recombinant vaccinia viruses, and colleagues at Covance Research Products, Inc. (Denver, PA) for conducting the rabbit immunization studies.
These studies were supported by HIVRAD grant P01 AI054564 (S.-L.H.), NICHD HD058304 (J.O.), and K08 AI068424 (C.A.B.).
Footnotes
Published ahead of print on 16 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddows, S., M. Franti, A. K. Dey, M. Kirschner, S. P. Iyer, D. C. Fisch, T. Ketas, E. Yuste, R. C. Desrosiers, P. J. Klasse, P. J. Maddon, W. C. Olson, and J. P. Moore. 2007. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360:329-340. [DOI] [PubMed] [Google Scholar]
- 3.Beddows, S., N. Schulke, M. Kirschner, K. Barnes, M. Franti, E. Michael, T. Ketas, R. W. Sanders, P. J. Maddon, W. C. Olson, and J. P. Moore. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 79:8812-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjouad, A., J. C. Gluckman, H. Rochat, L. Montagnier, and E. Bahraoui. 1992. Influence of carbohydrate moieties on the immunogenicity of human immunodeficiency virus type 1 recombinant gp160. J. Virol. 66:2473-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blish, C., R. Nedellec, K. Mandaliya, D. Mosier, and J. Overbaugh. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693-702. [DOI] [PubMed] [Google Scholar]
- 7.Blish, C. A., O. C. Dogan, N. R. Derby, M. A. Nguyen, B. Chohan, B. A. Richardson, and J. Overbaugh. 2008. HIV-1 superinfection occurs despite relatively robust neutralizing antibody responses. J. Virol. 82:12094-12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blish, C. A., Z. Jalalian-Lechak, S. Rainwater, M. A. Nguyen, O. C. Dogan, and J. Overbaugh. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D HIV-1 envelope variants. J. Virol. 83:7783-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blish, C. A., M. A. Nguyen, and J. Overbaugh. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolmstedt, A., J. Hinkula, E. Rowcliffe, M. Biller, B. Wahren, and S. Olofsson. 2001. Enhanced immunogenicity of a human immunodeficiency virus type 1 env DNA vaccine by manipulating N-glycosylation signals. Effects of elimination of the V3 N306 glycan. Vaccine 20:397-405. [DOI] [PubMed] [Google Scholar]
- 11.Bolmstedt, A., S. Sjolander, J. E. Hansen, L. Akerblom, A. Hemming, S. L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 12.Buonaguro, L., M. L. Tornesello, and F. M. Buonaguro. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 81:10209-10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 14.Burke, B., N. R. Derby, Z. Kraft, C. J. Saunders, C. Dai, N. Llewellyn, I. Zharkikh, L. Vojtech, T. Zhu, I. K. Srivastava, S. W. Barnett, and L. Stamatatos. 2006. Viral evolution in macaques coinfected with CCR5- and CXCR4-tropic SHIVs in the presence or absence of vaccine-elicited anti-CCR5 SHIV neutralizing antibodies. Virology 355:138-151. [DOI] [PubMed] [Google Scholar]
- 15.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Center, R. J., J. Lebowitz, R. D. Leapman, and B. Moss. 2004. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J. Virol. 78:2265-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti, S., J. R. Sisler, and B. Moss. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23:1094-1097. [DOI] [PubMed] [Google Scholar]
- 18.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 19:3979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ching, L. K., G. Vlachogiannis, K. A. Bosch, and L. Stamatatos. 2008. The first hypervariable region of the gp120 Env glycoprotein defines the neutralizing susceptibility of heterologous human immunodeficiency virus type 1 isolates to neutralizing antibodies elicited by the SF162gp140 immunogen. J. Virol. 82:949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derby, N. R., Z. Kraft, E. Kan, E. T. Crooks, S. W. Barnett, I. K. Srivastava, J. M. Binley, and L. Stamatatos. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 80:8745-8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey, B., M. Pancera, K. Svehla, Y. Shu, S. H. Xiang, J. Vainshtein, Y. Li, J. Sodroski, P. D. Kwong, J. R. Mascola, and R. Wyatt. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J. Virol. 81:5579-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose, N. A., G. H. Learn, A. G. Rodrigo, D. C. Nickle, F. Li, M. Mahalanabis, M. T. Hensel, S. McLaughlin, P. F. Edmonson, D. Montefiori, S. W. Barnett, N. L. Haigwood, and J. I. Mullins. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79:11214-11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl, P. L., and B. Moss. 1993. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res. Hum. Retroviruses 9:589-594. [DOI] [PubMed] [Google Scholar]
- 24.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsell, M. N., B. Dey, A. Morner, K. Svehla, S. O'Dell, C. M. Hogerkorp, G. Voss, R. Thorstensson, G. M. Shaw, J. R. Mascola, G. B. Karlsson Hedestam, and R. T. Wyatt. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4:e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnanakaran, S., D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn, and B. Korber. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 81:4886-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 30.Haigwood, N. L., P. L. Nara, E. Brooks, G. A. Van-Nest, G. Ott, K. W. Higgins, N. Dunlop, C. J. Scandella, J. W. Eichberg, and K. S. Steimer. 1992. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J. Virol. 66:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haigwood, N. L., J. R. Shuster, G. K. Moore, H. Lee, P. V. Skiles, K. W. Higgins, P. J. Barr, C. George-Nascimento, and K. S. Steimer. 1990. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res. Hum. Retroviruses 6:855-869. [DOI] [PubMed] [Google Scholar]
- 32.Hu, S. L., S. G. Kosowski, and J. M. Dalrymple. 1986. Expression of AIDS virus envelope gene in recombinant vaccinia viruses. Nature 320:537-540. [DOI] [PubMed] [Google Scholar]
- 33.Hu, S. L., and L. Stamatatos. 2007. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr. HIV Res. 5:507-513. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kothe, D. L., J. M. Decker, Y. Li, Z. Weng, F. Bibollet-Ruche, K. P. Zammit, M. G. Salazar, Y. Chen, J. F. Salazar-Gonzalez, Z. Moldoveanu, J. Mestecky, F. Gao, B. F. Haynes, G. M. Shaw, M. Muldoon, B. T. Korber, and B. H. Hahn. 2007. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kothe, D. L., Y. Li, J. M. Decker, F. Bibollet-Ruche, K. P. Zammit, M. G. Salazar, Y. Chen, Z. Weng, E. A. Weaver, F. Gao, B. F. Haynes, G. M. Shaw, B. T. Korber, and B. H. Hahn. 2006. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352:438-449. [DOI] [PubMed] [Google Scholar]
- 38.Kraft, Z., K. Strouss, W. F. Sutton, B. Cleveland, F. Y. Tso, P. Polacino, J. Overbaugh, S. L. Hu, and L. Stamatatos. 2008. Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses. J. Virol. 82:5912-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law, M., R. M. Cardoso, I. A. Wilson, and D. R. Burton. 2007. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J. Virol. 81:4272-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 80:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 47.Liao, H. X., L. L. Sutherland, S. M. Xia, M. E. Brock, R. M. Scearce, S. Vanleeuwen, S. M. Alam, M. McAdams, E. A. Weaver, Z. Camacho, B. J. Ma, Y. Li, J. M. Decker, G. J. Nabel, D. C. Montefiori, B. H. Hahn, B. T. Korber, F. Gao, and B. F. Haynes. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long, E. M., S. M. J. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retroviruses 18:567-576. [DOI] [PubMed] [Google Scholar]
- 49.Martin, H. L., D. J. Jackson, K. Mandaliya, J. Bwayo, J. P. Rakwar, P. Nyange, S. Moses, J. O. Ndinya-Achola, K. Holmes, F. Plummer, E. Ngugi, and J. Kreiss. 1994. Preparation for AIDS vaccine evaluation in Mombasa, Kenya: establishment of seronegative cohorts of commercial sex workers and trucking company employees. AIDS Res. Hum. Retroviruses 10:S235-S237. [PubMed] [Google Scholar]
- 50.Martin, H. L., P. M. Nyange, B. A. Richardson, L. Lavreys, K. Mandaliya, D. J. Jackson, J. O. Ndinya-Achola, and J. Kreiss. 1998. Hormonal contraception, sexually transmitted diseases, and the risk of heterosexual transmission of HIV-1. J. Infect. Dis. 178:1053-1059. [DOI] [PubMed] [Google Scholar]
- 51.Martin, H. L., B. A. Richardson, P. M. Nyange, L. Lavreys, S. L. Hillier, B. Chohan, K. Mandaliya, J. O. Ndinya-Achola, J. Bwayo, and J. Kreiss. 1999. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J. Infect. Dis. 180:1863-1868. [DOI] [PubMed] [Google Scholar]
- 52.Mascola, J. R., P. D'Souza, P. Gilbert, B. H. Hahn, N. L. Haigwood, L. Morris, C. J. Petropoulos, V. R. Polonis, M. Sarzotti, and D. C. Montefiori. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 54.Montefiori, D. 2004. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays, p. 12.11.1-12.11.17. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico (ed.), Current protocols in immunology, vol. 12. John Wiley & Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- 55.Quiñones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 57.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma, V. A., E. Kan, Y. Sun, Y. Lian, J. Cisto, V. Frasca, S. Hilt, L. Stamatatos, J. J. Donnelly, J. B. Ulmer, S. W. Barnett, and I. K. Srivastava. 2006. Structural characteristics correlate with immune responses induced by HIV envelope glycoprotein vaccines. Virology 352:131-144. [DOI] [PubMed] [Google Scholar]
- 59.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, D. C. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, A. Fong, S. R. Coates, L. Leung, M. Wininger, J. J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, G. Miiro, J. Serwanga, A. Pozniak, D. McPhee, O. Manigart, L. Mwananyanda, E. Karita, A. Inwoley, W. Jaoko, J. Dehovitz, L. G. Bekker, P. Pitisuttithum, R. Paris, S. Allen, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, X., A. B. Parast, B. A. Richardson, R. Nduati, G. John-Stewart, D. Mbori-Ngacha, S. M. Rainwater, and J. Overbaugh. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, Z. Y., B. K. Chakrabarti, L. Xu, B. Welcher, W. P. Kong, K. Leung, A. Panet, J. R. Mascola, and G. J. Nabel. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, P. F., F. Cham, M. Dong, A. Choudhary, P. Bouma, Z. Zhang, Y. Shao, Y. R. Feng, L. Wang, N. Mathy, G. Voss, C. C. Broder, and G. V. Quinnan, Jr. 2007. Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc. Natl. Acad. Sci. U. S. A. 104:10193-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.