Abstract
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene together represent the most common genetic determinant of Parkinson's disease (PD) identified to date. The vast majority of patients with LRRK2-related PD reported in the literature carry one of three pathogenic substitutions: G2019S, R1441C, or R1441G. While G2019S and R1441C are geographically widespread, R1441G is most prevalent in the Basque Country and is rare outside of Northern Spain. We sought to better understand the processes that have shaped the current distribution of R1441G. We performed a haplotype analysis of 29 unrelated PD patients heterozygous for R1441G and 85 wild-type controls using 20 markers that spanned 15.1 Mb across the LRRK2 region. Nine of the patients were of Basque origin and 20 were non-Basques. We inferred haplotypes using a Bayesian approach and utilized a maximum-likelihood method to estimate the age of the most recent common ancestor. Significant but incomplete allele sharing was observed over a distance of 6.0 Mb and a single, rare ten-marker haplotype 5.8 Mb in length was seen in all mutation carriers. We estimate that the most recent common ancestor lived 1,350 (95% CI, 1,020–1,740) years ago in approximately the seventh century. We hypothesize that R1441G originated in the Basque population and that dispersion of the mutation then occurred through short-range gene flow that was largely limited to nearby regions in Spain.
Keywords: Parkinson disease, LRRK2, R1441G, Founder effect, Basques
Introduction
Parkinson's disease (PD; MIM 168600) is the second most common neurodegenerative disorder and affects approximately 1–2% of the population over 60 years of age. It is characterized clinically by bradykinesia, resting tremor, rigidity, and postural instability, and pathologically by loss of dopamine neurons in the substantia nigra and Lewy body formation [1–3]. Only one in five PD patients report a family history of the disease and in most instances, PD is thought to result from a complex interaction between genetic and environmental factors [4, 5]. However, studies of rare multigenerational pedigrees in which PD segregates in a Mendelian pattern have yielded five “causal” genes: PARK2 (MIM 600116 and 602544), PINK1 (MIM 605909), PARK7 (MIM 606324), SNCA (MIM 163890), and LRRK2 (MIM 609007). Of these five genes, mutations in LRRK2 are the most prevalent in PD patients of European origin [6].
The majority of patients with LRRK2-related PD reported in the literature carry pathogenic variants within one of two mutational hotspots: codon 1441 in exon 31 (R1441C/G/H), and codon 2019 in exon 41 (G2019S). R1441C, R1441H, and G2019S have each arisen from at least three separate founding events and are widely geographically distributed [7–10]. All three mutations occur in Asians and in multiple European subpopulations. In contrast, R1441G is largely limited to Northern Spain. Originally, discovered in four families in the Basque Country [11], R1441G is found in approximately 20% of Basque patients with familial PD [12]. It was also identified at lower frequencies in patients from nearby provinces in Spain who did not report Basque ancestry [13, 14]. Previous work suggests that these patients might share the same background haplotype, but interpretation of these data is limited by a lack of overlap in the markers analyzed across studies [11, 12, 14, 15]. In this study, we sought to further explore whether PD patients who carry R1441G share a common founder, and if so to estimate the age of the founding event.
Materials and methods
Study participants
The study population was comprised of 29 unrelated PD patients who carried R1441G (Table 1), nine relatives of the patients, and 85 healthy mutation-negative controls from Northern Spain. Limited haplotype data on 14 of these patients have been published elsewhere [12, 14]. Twenty-eight of the patients were recruited from hospitals in three neighboring regions of Northern Spain (Asturias, n=15; Cantabria, n=1; Basque Country, n=12) and one (a Hispanic patient who did not report Basque ancestry) was ascertained from a movement disorder clinic in North America [16]. All patients met UK Parkinson's Disease Society Brain Bank clinical diagnostic criteria for PD [17]. The nine relatives came from three families (PJ68, FAM6, and FAM8); eight were affected and one was unaffected.
Table 1.
Characteristics of R1441G carriers
| No. of subjects | 29 |
|---|---|
| Age (mean±SD) | 62.4±11.3 |
| Age at onset (mean±SD) | 52.0±13.0 |
| Disease duration (mean±SD) | 11.8±7.1 |
| Family history of PD (%) | 65.5 |
| Male, n (%) | 15 (51.7) |
In Spain, an individual carries both of their parent's surnames throughout life and their name does not change with marriage. Thus, a great deal of information about recent ancestry can be gained by simply examining an individual's name. PD patients were classified as “Basque” if they had one or two Basque surnames and “non-Basque” if neither of their surnames was of Basque origin. Among the 29 PD patients included in the study, nine were categorized as Basque, and of these, four (FAM1, FAM3, FAM5, and FAM9) possessed two Basque surnames.
The controls were derived from two sources. Forty-two of the controls were autochthonous individuals from the Basque Country whose parents each had two Basque surnames (referred to hereafter as “Basque Controls”; mean age, 36.1±10.8 years; age range, 28–73 years; male, 47.6%). The remaining controls (n=43) were blood donors at one of two hospitals in Asturias, Spain (mean age, 40.9±11.5 years; age range, 20–62 years; male, 71.4%). Ancestral classification by surname was not possible for these individuals because data were collected anonymously.
The study was approved by the local ethics authorities at each institution and written informed consent was obtained from all participants.
Marker selection and genotyping
We selected a total of 20 markers (15 microsatellites and five single nucleotide polymorphisms [SNPs]) spanning a distance of 15.1 Mb across the LRRK2 region for genotyping in all study participants. We began with a set of 15 markers which have been used in several previous haplotype analyses of LRRK2 [11, 12, 14, 15]. We then added two microsatellites (D12S345 and D12S1713) and a SNP (rs1511547) chosen from the MAP-O-MAT database (http://compgen.rutgers.edu/mapomat/) to fill large gaps between markers. In genotyping rs1511547 (by sequencing) we identified two novel SNPs (rs55917927 and rs56260627) for which the minor allele was rare (frequency <13%) among controls but present in all patients carrying R1441G. These two potentially informative SNPs were also added to the marker set.
SNP genotyping was performed by sequencing with the Big-Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Microsatellites were amplified by PCR using fluorescently labelled forward primers. Genotypes were determined using an ABI PRISM 3130 Genetic Analyzer and GeneMapper 4.0 software (Applied Biosystems). Centre d'Etude du Polymorphisme Humain (CEPH) samples 1331-01, 1331-02, 1347-13, 1362-14, 1413-18, and 1416-12 (Coriell Cell Repositories, Camden NJ, USA) were used as a reference for microsatellite allele size determination. All PCR primer sequences and assay conditions are available on request.
Data analysis
We used PHASE software v2.1.1 (http://www.stat.washington.edu/stephens/software.html) to infer haplotypes when marker phase could not be resolved by pedigree data [18]. We estimated the age of the most recent common ancestor for individuals who carried R1441G using the program Estiage [19]. This maximum-likelihood algorithm uses information on the recombination fractions between the mutation and each marker, the frequencies of the shared allele at each marker, and the position of the first marker in each direction that is no longer shared, to calculate the number of generations (with 95% CI) elapsed since the most recent common ancestor introduced the mutation into the population. We defined a marker as shared if a single allele was included in all disease haplotypes or in at least half and at significantly greater frequency (Fisher's exact test) than in 170 inferred control haplotypes (collapsing all other alleles into a single bin). Genetic map positions for each marker were derived from the linkage-mapping server MAP-O-MAT, and physical positions were taken from the National Center for Biotechnology Information (NCBI) human genome assembly Build 36 [20].
Results
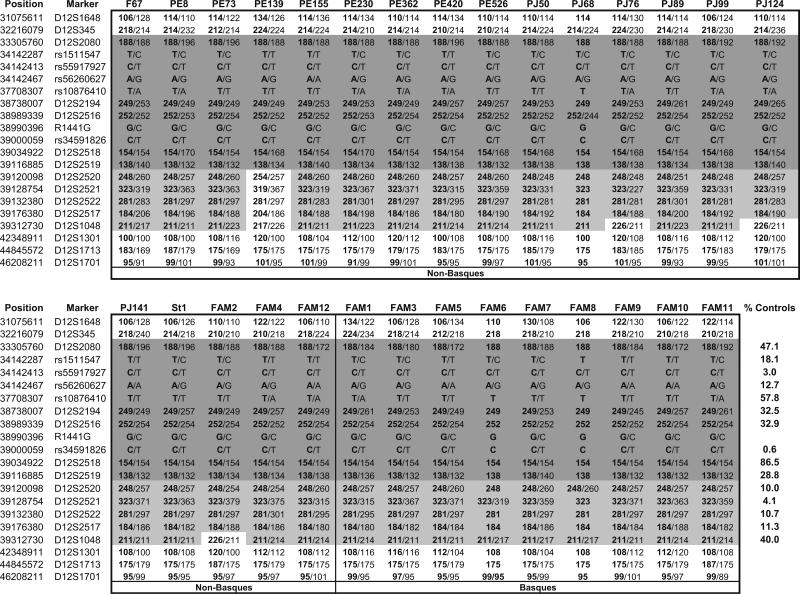
All 29 R1441G carriers shared a ten-marker background haplotype, bounded by D12S2080 and D12S2519, which spanned a distance of 5.8 Mb across the LRRK2 region (Fig. 1). The haplotype included alleles at two markers, rs55917927 and rs34591826 (M1646T), that were present at a frequency of only 3.0% and 0.6%, respectively, in control subjects. This haplotype was not observed among 170 control chromosomes. This strongly suggests that the mutation carriers in our study population originated from a single founder.
Fig. 1.
Genotypes for 20 markers surrounding the LRRK2 R1441G mutation. A single ten-marker haplotype (indicated by dark gray shading) was shared by all 29 mutation carriers. Incomplete but significant allele sharing is denoted by light gray shading and the frequency of each shared allele in 170 control chromosomes is displayedinthe lower right-hand margin. In instances in which phase could not be unambiguously determined, both alleles are shown. Disease haplotypes inferred using PHASE are indicated in bold type
The telomeric boundary of the haplotype shared by all mutation carriers was delimited by a single subject (PE139). In this individual, both alleles at D12S2520 and D12S2521 were divergent from the ones shared in all other carriers (Fig. 1). This suggests a previous recombination event between D12S2519 and D12S2520 for the disease chromosome carried by PE139, rather than recurrent mutation at two consecutive markers or an error in assigning phase. Among the disease chromosomes analyzed in the study, incomplete but significant allele sharing was seen across an interval of 6.0 Mb, from D12S2080 to D12S1048 (Fig. 1).
Because many of the markers in our dataset were tightly linked, genetic distances between them could not be accurately estimated from available genetic maps. Instead, we used the genetic length and physical distance between D12S2080 and D12S1301 to calculate an average of 0.30 cM/Mb across the entire region and then computed recombination fractions using the Kosambi mapping function as previously described [9]. We then estimated the age of the most recent common ancestor, designating D12S345 and D12S1301 as the first unshared markers in each direction. This yielded an age estimate of 45 (95% CI, 34–58) generations. Using the most widely published intergenerational interval, 25 years [21], our data indicate that the patients in our study shared a common ancestor 1,125 (95% CI, 850–1,450) years ago. However, several recent studies have concluded that 30 years is a better approximation of generation time for the period of human history in question [21–23]. Substituting a 30-year interval increases the age of the founding event to 1,350 (95% CI, 1,020–1,740) years ago. Using only Basque controls in the analysis yielded a very similar age estimate of 48 (95% CI, 37–63) generations. Excluding the subject (PE139) with the shortest shared haplotype also had little effect on the estimated age (45 generations; 95% CI, 35–58).
Discussion
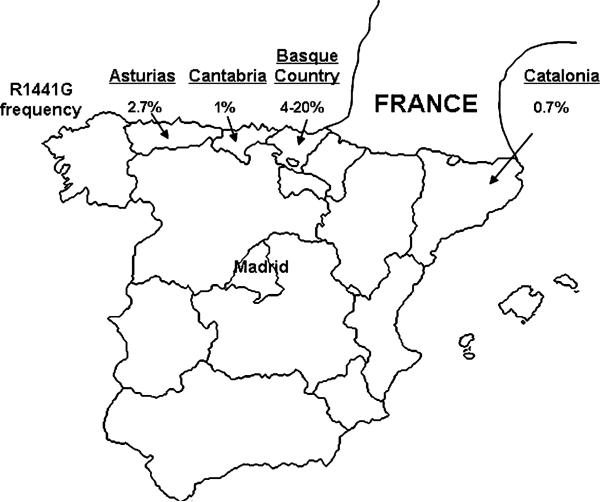
R1441G displays a frequency gradient which peaks in the Basque Country (Fig. 2) and it appears to be very rare outside of Northern Spain. Among several thousand PD patients from elsewhere in Europe and North America who have been screened for R1441G, only one (patient F67) has been found to carry the mutation [13, 16, 24–28]. Based on this pattern of distribution and the results of the present study, we hypothesize that R1441G originated in the Basque population and that a founding event occurred in approximately the seventh century. Dispersion of the mutation then occurred through short-range gene flow, largely limited to nearby regions. This scenario is consistent with current knowledge on the origin and history of the Basque population. The Basques are unusual among modern inhabitants of Europe in that they are thought to constitute a “relic” population descended from Paleolithic Europeans and have remained relatively genetically isolated [29, 30]. This view is supported by archaeological findings, linguistic studies, and analyses of both classic genetic and molecular markers [31–33]. However, examination of Y-chromosome markers also indicates that modest levels of gene flow have occurred between the Basques and neighboring populations (e.g. Catalans) over the past two millennia [34]. In addition, the distribution of mutations for other diseases that are believed to have originated among the Basques is similar to that seen for R1441G. This includes the CAPN3 2362AG→TCATCT mutation for limb-girdle muscular dystrophy type 2A (estimated to have arisen in the sixth to eighth century) [35] and PRNP D178N which results in fatal familial insomnia [36].
Fig. 2.
R1441G distribution in Spain. Mutation carrier frequencies based on previous studies [12–14] are shown for four different regions in Northern Spain
In contrast to R1441G, the LRRK2 G2019S mutation has been frequently observed across Europe, the Middle East, North Africa, and the Americas [12, 13, 37–43]. Most G2019S carriers share a common ancestor who is estimated to have lived approximately 2,250 years ago and likely originated in the Middle East [9]. While the spread of R1441G was probably slowed by the geographic and cultural boundaries that surround the Basque region, G2019S became widely dispersed, perhaps as a result of the large-scale migrations of the Jewish Diaspora.
Paisan-Ruiz and colleagues performed a haplotype analysis of the LRRK2 region in four extended Basque PD pedigrees with R1441G using a large set of SNP and microsatellite markers [11]. The authors later genotyped a subset of these markers (11 SNPs) that spanned 2.0 Mb across the region in 17 singleton PD cases (16 Basque and one non-Basque) who carried the mutation [15]. In both studies, they concluded that all mutation carriers shared a common founder, but did not calculate an age for the founding event. In the four Basque families and in the singleton group, complete allele sharing was observed over distances of 1.0 Mb and 170 kb, respectively. In our sample of 29 carriers, we observed complete allele sharing over a much larger distance of 5.8 Mb. Though the authors did not formally test for significant but incomplete allele sharing, at a marker (rs10876410) 1.3 Mb upstream from the mutation, only nine of the 17 singleton cases shared alleles. In contrast, in our sample we observed complete allele sharing at rs10876410 and at four markers further upstream, including D12S2080 located 5.7 Mb away from R1441G (Fig. 1). The shorter haplotypes observed by Paisan-Ruiz and colleagues imply that the founding event for R1441G might be substantially older than the estimate derived from our dataset. The reasons for the differences between our data and theirs are not entirely clear, and direct comparisons between datasets are difficult because only one of the markers (rs10876410) overlapped between studies. One possible reason is the nature of the markers used. While microsatellites are on average more informative, SNPs have much lower and less variable mutation rates [44]. Thus, within the same region, shared haplotypes based solely on microsatellites might be expected to be shorter than those based on SNPs alone. However, large differences in haplotype size remain even if one considers only the SNP markers in each dataset. Another consideration is whether the subjects in one or more studies might have actually arisen from multiple founders. In our sample, we believe that this is highly unlikely because all mutation carriers were heterozygous for rs34591826 (located 9.7 kb downstream of R1441G) and the minor allele frequency for this SNP was only 0.6% in controls (Fig. 1). The minor allele of rs34591826 was also included within the disease haplotype in the four large Basque families [11]. However, in the 17 singleton PD cases rs34591826 was not genotyped and the four SNPs (rs4768224, rs12423567, rs1820544, and rs10784616) that constituted the core haplotype shared by all subjects in that study were less informative [15]. The shared alleles for these four SNPs had frequencies ranging from 0.28 to 0.70 in the HapMap CEU sample (http://www.hapmap.org). Thus, there is somewhat less certainty that all of the individuals in this singleton PD sample did indeed descend from a common ancestor, particularly because the analysis was conducted with phase-unknown data and statistical methods were not used to infer phase. Finally, another possibility to explain the differences between our data and those of Paisan-Ruiz and colleagues is genotyping error.
Our study also had some limitations. Because DNA from family members was available for only three mutation carriers, we used Bayesian methods to reconstruct haplotypes in most instances. This has the potential to introduce additional uncertainty into our estimates of the age of the founding event which is not taken into account with the maximum-likelihood methods we used. Also, the use of surnames to categorize mutation carriers as Basque or non-Basque is subject to potential misclassification and it is possible that the ancestors of some subjects came from both groups.
Our data provide further empirical evidence that the Basques have remained genetically isolated over the past one to two millennia. Our findings also lend further support to the idea that important genetic determinants for PD can be highly population-specific. Finally, there is little information available on the prevalence of R1441G in Central and South America where the majority of intercontinental Basque migration has occurred. We are addressing this issue in ongoing studies of the LRRK2 gene in PD cohorts from across these regions.
Acknowledgements
We dedicate this paper to our dear colleague, Dr. Luis M. Guisasola, a superb clinician, researcher, and educator, who recently passed away. We thank the individuals who participated in the study. This work was supported by the Basque Government and University of the Basque Country (grant S-PE07UN44, M.M.P. and M.C.G-F.); the NIH (NINDS, K08 NS044138, C.P.Z.); the Department of Veterans Affairs (Merit Review Award, C.P.Z.); the Parkinson's Disease Foundation (Fellowship Award, I.F.M.); the Spanish Fondo de Investigacion Sanitaria (grant FIS PI070014, J.I; grant 05/008, V.A.); and the Veterans Integrated Service Network 20 Geriatric, Mental Illness, and Parkinson's Disease Research, Education, and Clinical Centers.
Contributor Information
Ignacio F. Mata, Geriatric Research Education and Clinical Center S-182, Veterans Affairs Puget Sound Health Care System, 1660 South Columbian Way, Seattle, WA 98108, USA Department of Neurology, University of Washington, Seattle, WA, USA.
Carolyn M. Hutter, Department of Epidemiology, University of Washington, Seattle, WA, USA
María C. González-Fernández, Servicio General de Investigación Genómica: Banco de ADN, Universidad del País Vasco, Vitoria-Gasteiz, Spain
Marian M. de Pancorbo, Servicio General de Investigación Genómica: Banco de ADN, Universidad del País Vasco, Vitoria-Gasteiz, Spain
Elena Lezcano, Unidad de trastornos del movimiento, Hospital de Cruces, Baracaldo, Spain.
Cecilia Huerta, Genética Molecular-Instituto de Investigacion Nefrológica, Hospital Universitario Central de Asturias, Oviedo, Spain.
Marta Blazquez, Servicio de Neurología, Hospital Universitario Central de Asturias, Oviedo, Spain.
Renee Ribacoba, Servicio de Neurología, Hospital Alvarez-Buylla, Mieres, Spain.
Luis M. Guisasola, Servicio de Neurología, Hospital Universitario Central de Asturias, Oviedo, Spain
Carlos Salvador, Servicio de Neurología, Hospital Universitario Central de Asturias, Oviedo, Spain.
Juan C. Gómez-Esteban, Unidad de trastornos del movimiento, Hospital de Cruces, Baracaldo, Spain
Juan J. Zarranz, Unidad de trastornos del movimiento, Hospital de Cruces, Baracaldo, Spain
Jon Infante, Servicio de Neurología, Hospital Universitario “Marqués de Valdecilla”, Universidad de Cantabria, Santander, Spain.
Joseph Jankovic, Department of Neurology, Baylor College of Medicine, Houston, TX, USA.
Hao Deng, Department of Neurology, Baylor College of Medicine, Houston, TX, USA; Center for Experimental Medicine, The Third Xiangya Hospital, Central South University, Changsha, China.
Karen L. Edwards, Department of Epidemiology, University of Washington, Seattle, WA, USA
Victoria Alvarez, Genética Molecular-Instituto de Investigacion Nefrológica, Hospital Universitario Central de Asturias, Oviedo, Spain.
Cyrus P. Zabetian, Geriatric Research Education and Clinical Center S-182, Veterans Affairs Puget Sound Health Care System, 1660 South Columbian Way, Seattle, WA 98108, USA zabetian@u.washington.edu Department of Neurology, University of Washington, Seattle, WA, USA.
References
- 1.Samii A, Nutt JG, Ransom BR. Parkinson's disease. Lancet. 2004;363:1783–1793. doi: 10.1016/S0140-6736(04)16305-8. doi:10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 2.Przedborski S, Tieu K, Perier C, Vila M. MPTP as a mitochondrial neurotoxic model of Parkinson's disease. J Bioenerg Biomembr. 2004;36:375–379. doi: 10.1023/B:JOBB.0000041771.66775.d5. doi:10.1023/B:JOBB.0000041771.66775.d5. [DOI] [PubMed] [Google Scholar]
- 3.Jankovic J. Parkinson's disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. doi:10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 4.Payami H, Larsen K, Bernard S, Nutt J. Increased risk of Parkinson's disease in parents and siblings of patients. Ann Neurol. 1994;36:659–661. doi: 10.1002/ana.410360417. doi:10.1002/ana.410360417. [DOI] [PubMed] [Google Scholar]
- 5.Bonifati V, Fabrizio E, Vanacore N, De Mari M, Meco G. Familial Parkinson's disease: a clinical genetic analysis. Can J Neurol Sci. 1995;22:272–279. doi: 10.1017/s0317167100039469. [DOI] [PubMed] [Google Scholar]
- 6.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. doi:10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 7.Haugarvoll K, Rademakers R, Kachergus JM, Nuytemans K, Ross OA, Gibson JM, Tan EK, Gaig C, Tolosa E, Goldwurm S, Guidi M, Riboldazzi G, Brown L, Walter U, Benecke R, Berg D, Gasser T, Theuns J, Pals P, Cras P, De Deyn PP, Engelborghs S, Pickut B, Uitti RJ, Foroud T, Nichols WC, Hagenah J, Klein C, Samii A, Zabetian CP, Bonifati V, Van Broeckhoven C, Farrer MJ, Wszolek ZK. Lrrk2 R1441C parkinsonism is clinically similar to sporadic Parkinson disease. Neurology. 2008;70:1456–1460. doi: 10.1212/01.wnl.0000304044.22253.03. doi:10.1212/01.wnl.0000304044.22253.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross OA, Spanaki C, Griffith A, Lin CH, Kachergus J, Haugarvoll K, Latsoudis H, Plaitakis A, Ferreira JJ, Sampaio C, Bonifati V, Wu RM, Zabetian CP, Farrer MJ. Haplotype analysis of Lrrk2 R1441H carriers with parkinsonism. Parkinsonism Relat Disord. 2008 doi: 10.1016/j.parkreldis.2008.09.001. doi:10.1016/j.parkreldis.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zabetian CP, Hutter CM, Yearout D, Lopez AN, Factor SA, Griffith A, Leis BC, Bird TD, Nutt JG, Higgins DS, Roberts JW, Kay DM, Edwards KL, Samii A, Payami H. LRRK2 G2019S in families with Parkinson Disease who originated from Europe and the Middle East: evidence of two distinct founding events beginning two Millennia ago. Am J Hum Genet. 2006;79:752–758. doi: 10.1086/508025. doi:10.1086/508025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zabetian CP, Morino H, Ujike H, Yamamoto M, Oda M, Maruyama H, Izumi Y, Kaji R, Griffith A, Leis BC, Roberts JW, Yearout D, Samii A, Kawakami H. Identification and haplotype analysis of LRRK2 G2019S in Japanese patients with Parkinson disease. Neurology. 2006;67:697–99. doi: 10.1212/01.wnl.0000227732.37801.d4. doi:10.1212/01.wnl.0000227732.37801.d4. [DOI] [PubMed] [Google Scholar]
- 11.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, de Munain AL, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. doi:10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Fernandez MC, Lezcano E, Ross OA, Gomez-Esteban JC, Gomez-Busto F, Velasco F, Alvarez-Alvarez M, Rodriguez-Martinez MB, Ciordia R, Zarranz JJ, Farrer MJ, Mata IF, de Pancorbo MM. Lrrk2-associated parkinsonism is a major cause of disease in Northern Spain. Parkinsonism Relat Disord. 2007;13:509–515. doi: 10.1016/j.parkreldis.2007.04.003. doi:10.1016/j.parkreldis.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Gaig C, Ezquerra M, Marti MJ, Munoz E, Valldeoriola F, Tolosa E. LRRK2 mutations in Spanish patients with Parkinson disease: frequency, clinical features, and incomplete penetrance. Arch Neurol. 2006;63:377–382. doi: 10.1001/archneur.63.3.377. doi:10.1001/archneur.63.3.377. [DOI] [PubMed] [Google Scholar]
- 14.Mata IF, Taylor JP, Kachergus J, Hulihan M, Huerta C, Lahoz C, Blazquez M, Guisasola LM, Salvador C, Ribacoba R, Martinez C, Farrer M, Alvarez V. LRRK2 R1441G in Spanish patients with Parkinson's disease. Neurosci Lett. 2005;382:309–311. doi: 10.1016/j.neulet.2005.03.033. doi:10.1016/j.neulet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Simon-Sanchez J, Marti-Masso JF, Sanchez-Mut JV, Paisan-Ruiz C, Martinez-Gil A, Ruiz-Martinez J, Saenz A, Singleton AB, Lopez de Munain A, Perez-Tur J. Parkinson's disease due to the R1441G mutation in Dardarin: a founder effect in the Basques. Mov Disord. 2006;21:1954–1959. doi: 10.1002/mds.21114. doi:10.1002/mds.21114. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Le W, Guo Y, Hunter CB, Xie W, Huang M, Jankovic J. Genetic analysis of LRRK2 mutations in patients with Parkinson disease. J Neurol Sci. 2006;251:102–106. doi: 10.1016/j.jns.2006.09.017. doi:10.1016/j.jns.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J Neurol Neurosurg Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. doi:10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. doi:10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–449. doi: 10.1136/jmg.2003.017962. doi:10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong X, Matise TC. MAP-O-MAT: internet-based linkage mapping. Bioinformatics. 2005;21:557–559. doi: 10.1093/bioinformatics/bti024. doi:10.1093/bioinformatics/bti024. [DOI] [PubMed] [Google Scholar]
- 21.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. doi:10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 22.Sigurgardottir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P. The mutation rate in the human mtDNA control region. Am J Hum Genet. 2000;66:1599–1609. doi: 10.1086/302902. doi:10.1086/302902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremblay M, Vezina H. New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet. 2000;66:651–658. doi: 10.1086/302770. doi:10.1086/302770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, Strain KJ, Maraganore DM. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. doi:10.1212/01.WNL.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 25.Goldwurm S, Di Fonzo A, Simons EJ, Rohe CF, Zini M, Canesi M, Tesei S, Zecchinelli A, Antonini A, Mariani C, Meucci N, Sacilotto G, Sironi F, Salani G, Ferreira J, Chien HF, Fabrizio E, Vanacore N, Dalla Libera A, Stocchi F, Diroma C, Lamberti P, Sampaio C, Meco G, Barbosa E, Bertoli-Avella AM, Breedveld GJ, Oostra BA, Pezzoli G, Bonifati V. The G6055A (G2019S) mutation in LRRK2 is frequent in both early and late onset Parkinson's disease and originates from a common ancestor. J Med Genet. 2005;42:e65. doi: 10.1136/jmg.2005.035568. doi:10.1136/jmg.2005.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols WC, Elsaesser VE, Pankratz N, Pauciulo MW, Marek DK, Halter CA, Rudolph A, Shults CW, Foroud T. LRRK2 mutation analysis in Parkinson disease families with evidence of linkage to PARK8. Neurology. 2007;69:1737–1744. doi: 10.1212/01.wnl.0000278115.50741.4e. doi:10.1212/01.wnl.0000278115.50741.4e. [DOI] [PubMed] [Google Scholar]
- 27.Berg D, Schweitzer K, Leitner P, Zimprich A, Lichtner P, Belcredi P, Brussel T, Schulte C, Maass S, Nagele T. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease*. Brain. 2005;128:3000–3011. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 28.Zabetian CP, Samii A, Mosley AD, Roberts JW, Leis BC, Yearout D, Raskind WH, Griffith A. A clinic-based study of the LRRK2 gene in Parkinson disease yields new mutations. Neurology. 2005;65:741–744. doi: 10.1212/01.wnl.0000172630.22804.73. doi:10.1212/01.WNL.0000172630.22804.73. [DOI] [PubMed] [Google Scholar]
- 29.Cavalli-Sforza LL, Piazza A. Human genomic diversity in Europe: a summary of recent research and prospects for the future. Eur J Hum Genet. 1993;1:3–18. doi: 10.1159/000472383. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez AM, Garcia O, Larruga JM, Cabrera VM. The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country. BMC Genomics. 2006;7:124. doi: 10.1186/1471-2164-7-124. doi:10.1186/1471-2164-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfonso-Sanchez MA, Cardoso S, Martinez-Bouzas C, Pena JA, Herrera RJ, Castro A, Fernandez-Fernandez I, De Pancorbo MM. Mitochondrial DNA haplogroup diversity in Basques: a reassessment based on HVI and HVII polymorphisms. Am J Hum Biol. 2008;20:154–164. doi: 10.1002/ajhb.20706. doi:10.1002/ajhb.20706. [DOI] [PubMed] [Google Scholar]
- 32.de Pancorbo MM, Lopez-Martinez M, Martinez-Bouzas C, Castro A, Fernandez-Fernandez I, de Mayolo GA, de Mayolo AA, de Mayolo PA, Rowold DJ, Herrera RJ. The Basques according to polymorphic Alu insertions. Hum Genet. 2001;109:224–233. doi: 10.1007/s004390100544. doi:10.1007/s004390100544. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Miranda AM, Alfonso-Sanchez MA, Kalantar A, Pena JA, Pancorbo MM, Herrera RJ. Allelic frequencies of 13 STR loci in autochthonous Basques from the province of Vizcaya (Spain). Forensic Sci Int. 2005;152:259–262. doi: 10.1016/j.forsciint.2004.09.118. doi:10.1016/j.forsciint.2004.09.118. [DOI] [PubMed] [Google Scholar]
- 34.Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Perez-Lezaun A, Bosch E, Shlumukova M, Cambon-Thomsen A, McElreavey K, Lopez De Munain A, Rohl A, Wilson IJ, Singh L, Pandya A, Santos FR, Tyler-Smith C, Jobling MA. Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet. 1999;65:1437–1448. doi: 10.1086/302617. doi:10.1086/302617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobo AM, Saenz A, Poza JJ, Urtasun M, Indakoetxea B, Urtizberea JA, Lopez de Munain A, Calafell F. A common haplotype associated with the Basque 2362AG –> TCATCT mutation in the muscular calpain-3 gene. Hum Biol. 2004;76:731–741. doi: 10.1353/hub.2005.0002. doi:10.1353/hub.2005.0002. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Martinez AB, Alfonso-Sanchez MA, Pena JA, Sanchez-Valle R, Zerr I, Capellari S, Calero M, Zarranz JJ, de Pancorbo MM. Molecular evidence of founder effects of fatal familial insomnia through SNP haplotypes around the D178N mutation. Neurogenetics. 2008;9:109–118. doi: 10.1007/s10048-008-0120-x. doi:110.1007/s10048-10008-10120-x. [DOI] [PubMed] [Google Scholar]
- 37.Kay DM, Zabetian CP, Factor SA, Nutt JG, Samii A, Griffith A, Bird TD, Kramer P, Higgins DS, Payami H. Parkinson's disease and LRRK2: frequency of a common mutation in U.S. movement disorder clinics. Mov Disord. 2006;21:519–523. doi: 10.1002/mds.20751. doi:10.1002/mds.20751. [DOI] [PubMed] [Google Scholar]
- 38.Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, Payami H, Nutt J, Maraganore DM, Czyzewski K, Styczynska M, Wszolek ZK, Farrer MJ, Toft M. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76:672–680. doi: 10.1086/429256. doi:10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, Lipton RB, Soto-Valencia J, Risch N, Bressman SB. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. doi:10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 40.Orr-Urtreger A, Shifrin C, Rozovski U, Rosner S, Bercovich D, Gurevich T, Yagev-More H, Bar-Shira A, Giladi N. The LRRK2 G2019S mutation in Ashkenazi Jews with Parkinson disease: is there a gender effect. Neurology. 2007;69:1595–1602. doi: 10.1212/01.wnl.0000277637.33328.d8. doi:10.1212/01.wnl.0000277637.33328.d8. [DOI] [PubMed] [Google Scholar]
- 41.Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. doi:10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 42.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 43.Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, Singleton A, Foroud T. Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 44.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. doi:10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]