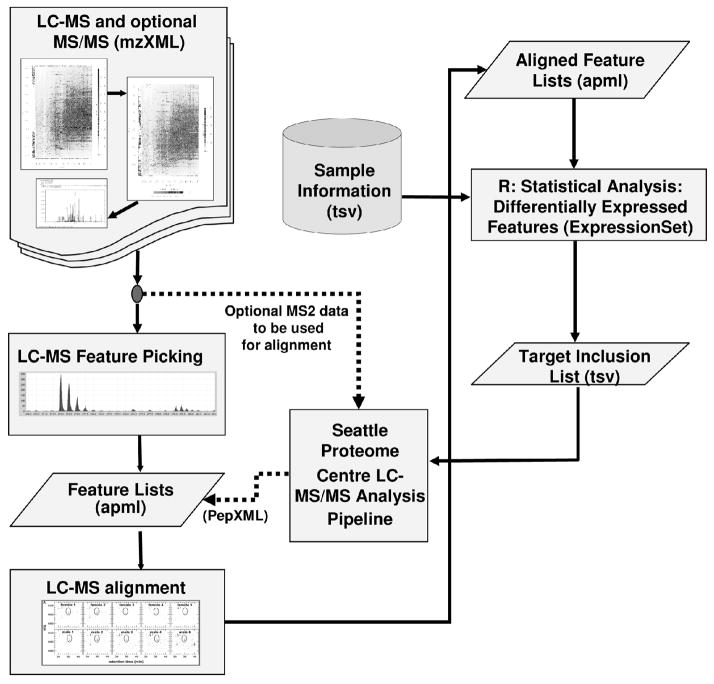
Figure 1.
Workflow of the Corra framework. The pipeline accepts mzXML files as input, then it performs peak picking and alignment. The statistics module is used to find differentially abundant features that are significant, and generates an inclusion list from them. Identified peptides are then annotated back to the feature list.