Figure 1.
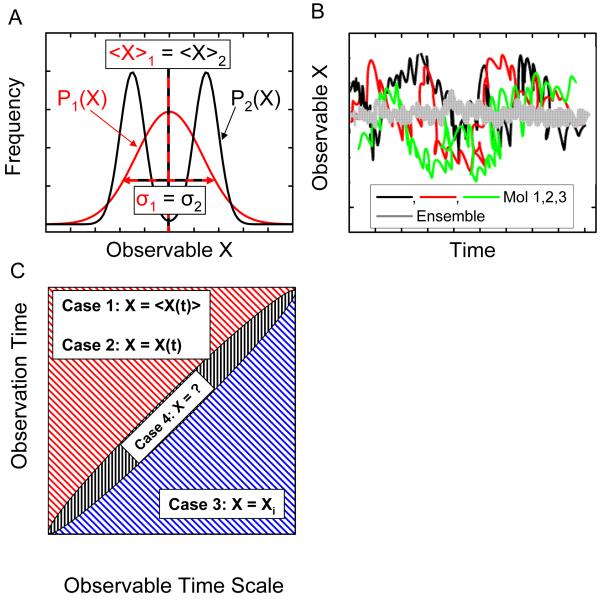
Single-molecule measurements give access to information hidden in ensemble measurements. A. Two distributions of an observable X are represented. The first distribution, P1(X), is a simple Gaussian corresponding for instance to a single species of molecule, or a single state. The second distribution, P2(X), is the sum of two Gaussians, corresponding to two different species, or two different possible states of a molecule. Both distributions have the same mean and standard deviation, and would result in indistinguishable ensemble measurements, whereas single-molecule experiments have the potential to give the complete distribution by separately observing molecules with different X values, revealing the static heterogeneity of sample 2. B. In a sample characterized by dynamic heterogeneity, molecules in an ensemble have non-synchronized evolutions, and any measurement will only give the average value of the observable. Single-molecule experiments, by giving access to the temporal evolution of individual molecule, or by allowing capturing snapshots of many different molecules, allow the full phase space to be recovered. C. The dynamic of single molecules can be captured if and only if the duration of the observation exceeds the typical time scale of the dynamic to be observed (upper left part of the graph). Two limit cases are possible. Case 1: the duration of the experiment is long enough to encompass different molecular states, but the experimental time resolution is not fast enough to capture them: the measured observable is a time average of the observable, <X(t)>, over the minimum integration time; Case 2: if the time resolution is sufficient, the measurement will give access to the full temporal dynamic, X(t), allowing to observe equilibrium fluctuations or non-equilibrium trajectories. If, on the other hand, the duration of the observation is shorter than the typical dynamic time scale, the measurement can only capture a snapshot, Xi, of the observable (case 3, lower right). In between these two regimes (case 4), the resolution will be too long to capture snapshots of the molecular state, and to short to properly average out the observable, resulting in hard to analyze fluctuating data