Abstract
Glutamate is a major excitatory neurotransmitter in the vertebrate brain. AMPA-type glutamate receptors mediate fast excitatory transmission. AMPA receptors assemble with transmembrane AMPA receptor regulatory protein (TARP) auxiliary subunits and function as native ion channels. However, the assembly and stoichiometry of AMPA receptor and TARP complexes remain unclear. Here, we developed a novel strategy to determine the assembly and stoichiometry of this protein complex and found that functional AMPA receptors indeed assembled as a tetramer in a dimer-of-dimers structure. Furthermore, we found that the AMPA receptor auxiliary subunit, TARP, had a variable stoichiometry (1–4 TARP units) on AMPA receptors and that 1 TARP unit was sufficient to modulate AMPA receptor activity. In neurons, TARP had fixed and minimum stoichiometry on AMPA receptors. This fundamental composition of the AMPA receptor/TARP complex is important for the elucidation of the molecular machinery that underlies synaptic transmission.
Introduction
Glutamate is a major excitatory neurotransmitter in the vertebrate brain, and AMPA-type glutamate receptors play critical roles in fast synaptic transmission. AMPA receptors have four subunits (GluA1–4) (Seeburg, 1993; Hollmann and Heinemann, 1994). Transmembrane AMPA receptor regulatory proteins (TARPs) have been identified as auxiliary subunits of AMPA receptors (Nicoll et al., 2006; Osten and Stern-Bach, 2006; Ziff, 2007; Coombs and Cull-Candy, 2009; Sager et al., 2009). AMPA receptors and their auxiliary subunits assemble and function as native ion channels in the brain.
The assembly and stoichiometry of AMPA receptors have been studied extensively. Recombinant AMPA receptors exhibit three distinct conductance levels in a single-channel recording (Rosenmund et al., 1998). Leucine zipper peptide-based oligomerization assays showed that tetramerized AMPA receptors work more efficiently than monomeric, dimeric, trimeric, and pentameric peptide-fused receptors (Matsuda et al., 2005). A chemical cross-linking experiment of native AMPA receptors from porcine brain revealed the presence of multiple bands; the molecular weight of the largest complex was ∼400 kDa (Wu et al., 1996). AMPA receptors were detected on blue native (BN)-PAGE predominantly as tetramers and weakly as monomers and dimers in neurons (Greger et al., 2002, 2003, 2006; Vandenberghe et al., 2005). In addition, sedimentation equilibrium analysis of the ligand-binding domains (S1-S2) of the AMPA receptor in solution revealed that these domains form a dimer after binding of cyclothiazide, which is a desensitization blocker of AMPA receptors (Sun et al., 2002). Single-particle analysis of AMPA receptors purified from rat brains and from SF9 cells revealed the presence of twofold asymmetric and symmetric structures, respectively (Safferling et al., 2001; Nakagawa et al., 2005; Midgett and Madden, 2008). The N-terminal domain (NTD) of the AMPA receptor can form a dimer (Leuschner and Hoch, 1999) independently of the ligand-binding domains. The crystal structure of this NTD was resolved recently and confirmed that the NTD forms a dimer (Jin et al., 2009; Kumar et al., 2009). In addition, the Q/R editing site in the pore loop of AMPA receptors was suggested to play a role in AMPA receptor tetramerization (Greger et al., 2003). These results strongly indicate that the AMPA receptor is a tetramer that forms a dimer-of-dimers structure. Consistent with the dimer-of-dimers model, the functional characterization of AMPA receptor mutants suggests that this receptor is a tetramer (Mano and Teichberg, 1998; Robert et al., 2001) and that the dimer-of-dimers model fits well with reported results (Ayalon and Stern-Bach, 2001; Mansour et al., 2001). However, the function of TARPs as AMPA receptor auxiliary subunits and the stoichiometry of TARPs are unknown.
Here, we developed a novel strategy based on SDS-PAGE and BN-PAGE to explore the assembly and stoichiometric properties of AMPA receptor and TARP complexes. We found that the functional AMPA receptor was a tetramer that indeed formed a dimer-of-dimers structure. TARPs showed a variable stoichiometry (1–4 units) on AMPA receptors, and each of the four TARP isoforms interacted with the AMPA receptor independently, without any cooperative binding properties. In neurons, TARP had fixed and minimum stoichiometry on AMPA receptors. This fundamental composition of the AMPA receptor/TARP complex is important for the elucidation of the molecular machinery that underlies synaptic transmission.
Materials and Methods
Antibodies.
The following antibodies were used: rabbit polyclonal antibodies to GluA1, GluA2/3, GluA4, and pan-TARP (Millipore); guinea pig polyclonal antibody to green florescent protein (GFP) (Tomita et al., 2004); and mouse monoclonal antibody to hemagglutinin (HA) epitope (Covance).
Plasmid construction.
GluA1 and stargazin (STG) were subcloned into pGEMHE with multiple units of AcGFP (Clontech).
Electrophysiology using Xenopus laevis oocytes.
Two-electrode voltage-clamp (TEVC) recordings were performed as described previously (Tomita et al., 2004). Briefly, cRNAs were transcribed in vitro using T7 mMessage mMachine (Ambion) and oocytes were injected with GluA1 cRNA alone or with GluA1 and stargazin cRNAs at the amount indicated. TEVC analysis (holding potential = −70 mV) was performed 2 d after injection at room temperature. Each agonist was bath applied in recording solution containing the following (in mm): 90 NaCl, 1.0 KCl, 1.5 CaCl2, and 10 HEPES, pH 7.4. Data were presented as mean ± SEM. Differences in means were tested using one-way ANOVA followed by post hoc analysis with Tukey's test.
BN-PAGE.
BN-PAGE was performed as described previously (Schagger et al., 1994) and gel concentrations are indicated in the legends of the figures. Oocytes were injected with GluA1 cRNA alone or with GluA1 and stargazin cRNAs at the concentrations indicated. Oocytes were homogenized in 20 mm Tris/5 mm EGTA pH8.0 using a Dounce homogenizer. After centrifugation at 20,000 × g for 20 s, the pellet was solubilized with 0.3% Triton X-100 for 30 min at 4°C, followed by centrifugation at 20,000 × g for 5 min. The solubilized proteins were then resolved by SDS-PAGE or BN-PAGE, which was followed by Western blot analysis. Films were scanned and the signal intensity of each band was analyzed using the ImageJ software, which is available from the NIH website, followed by normalization of signals to the wild-type signal after subtraction of the background signal of the film. Data were presented as means ± SEM. Differences in means were tested using one-way ANOVA, followed by post hoc analysis with Tukey's test, or Student's t test and are shown in each figure legend.
Agonist-evoked currents in cerebellar granule cells.
Stargazer mice were obtained from the Jackson Laboratory and were maintained at the Yale animal facility under the guidelines of the Institutional Animal Care and Use Committee. Heterozygous male and female mice were mated to obtain wild-type, heterozygous, and homozygous stargazer mice. Cerebellar granule cell cultures were prepared from postnatal day 7 mice. Patch-clamp recordings from cerebellar granule cells (DIV7–9) were performed in external solution, pH 7.4, containing the following (in mm): 10 HEPES, 140 NaCl, 2.5 KCl, 2.5 CaCl2, 1.3 MgSO4, 2.7 MgCl2, and 10 glucose. Patch pipettes were filled with a recording solution (pH 7.2, 320 mOsm) that contained the following (in mm): 130 cesium methanesulfonate, 5 HEPES, 5 Mg-ATP, 0.2 Na-GTP, 20 tetraethylammonium, and 5 EGTA. All recordings were performed at room temperature. To isolate and record AMPA receptor-mediated currents, tetrodotoxin (1 μm), AP-5 (100 μm), and picrotoxin (100 μm) were added to the external solution. The current was analog low-pass filtered at 3 kHz and digitally sampled at 25 kHz. Pipette resistances for these experiments were typically ∼3–5 MΩ and series resistances were ∼15–20 MΩ. Only recording epochs in which series and input resistances varied <10% were analyzed. Data were presented as mean ± SEM. Differences between experimental groups were considered significant when p was <0.05 by Tukey's test with ANOVA.
Results
The AMPA receptor assembles with TARP
AMPA receptors function as hetero-oligomers or homo-oligomers and TARPs function as AMPA-receptor auxiliary subunits. To determine the assembly and stoichiometry of the AMPA receptor/TARP complex, i.e., the specific ratio of molecules present in the functional AMPA receptor complex, we used BN-PAGE, which has the advantage of preserving protein complexes on PAGE (Schagger et al., 1994) (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material).
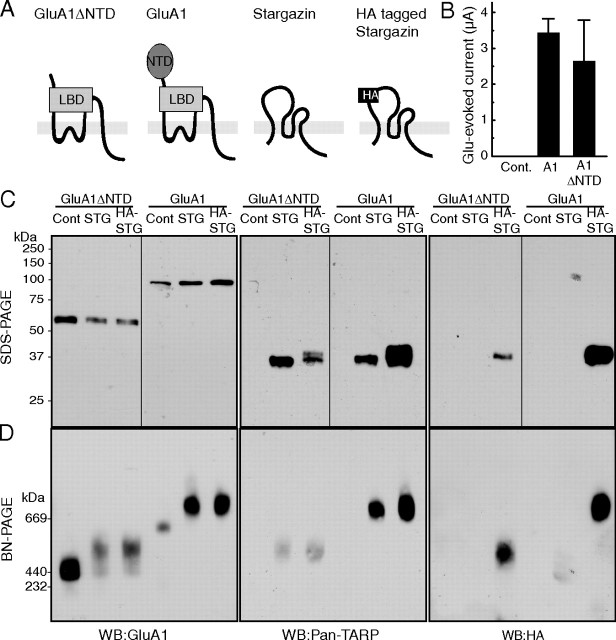
To detect the AMPA receptor/TARP complex using BN-PAGE, we selected the GluA1 subunit of the AMPA receptor and the prototypical TARP isoform stargazin/γ-2. We expressed GluA1 and GluA1 lacking the large NTD (GluA1ΔNTD) in Xenopus laevis oocytes via injection of their respective cRNAs in the presence or absence of stargazin or stargazin tagged with an HA epitope in the first extracellular loop (HA-stargazin) (Fig. 1A). We confirmed that both AMPA receptors used here exhibited comparable ion channel activity (Fig. 1B) (Tomita et al., 2004, 2007a). Expression of full-length proteins without protein degradation was confirmed by SDS-PAGE using an anti-GluA1 antibody (which recognizes the C terminus of GluA1), an anti-pan-TARP antibody (which recognizes the cytoplasmic domain of stargazin), and an anti-HA antibody (which recognizes the HA epitope inserted at the first extracellular loop of stargazin) (Fig. 1C). Stargazin was detected at ∼37 kDa and GluA1 and GluA1ΔNTD were detected as single bands that migrated at ∼100 kDa and ∼55 kDa, respectively (Fig. 1C).
Figure 1.
AMPA receptors assemble with TARP/stargazin. A, Schematic domain structure of GluA1, GluA1 lacking the NTD (GluA1ΔNTD), stargazin, and HA-epitope-tagged stargazin (HA-stargazin). LBD, Ligand-binding domain. B–D, Oocytes were injected with cRNA of GluA1 (10 ng), GluA1ΔNTD (5 ng), stargazin, and HA-stargazin (0.25 ng), which was followed by measuring glutamate (100 μm)-evoked current with cyclothiazide (50 μm) by two-electrode voltage-clamp recording (B) and solubilization with Triton X-100 and SDS-PAGE (10%) (C), or BN-PAGE (4–8%) (D). Proteins were detected using an anti-GluA1 antibody (which recognizes the C terminus of GluA1), an anti-pan-TARP antibody (which recognizes the cytoplasmic domain of stargazin), and an anti-HA-epitope antibody. C, GluA1, stargazin (STG), and HA-stargazin were expressed at 100, 37, and 37 kDa, respectively, without detection of protein degradation on SDS-PAGE. D, GluA1 and GluA1ΔNTD formed homo-oligomers around 669 and 440 kDa, respectively, on BN-PAGE. The molecular weight of GluA1 and GluA1ΔNTD homo-oligomers shifted toward a higher molecular weight after coexpression with stargazin or HA-stargazin. The shifted bands of GluA1 and GluA1ΔNTD homo-oligomers were also recognized by an anti-pan-TARP antibody and an anti-HA antibody. WB, Western blotting; Cont, water-injected control.
GluA1 and GluA1ΔNTD were detected as single bands that migrated on BN-PAGE at ∼669 kDa and ∼440 kDa, respectively (Fig. 1D). Coexpression of stargazin and HA-stargazin shifted the molecular weight of the GluA1 complex toward a higher molecular weight on BN-PAGE (Fig. 1D). The shifted band was also recognized by the anti-pan-TARP and anti-HA antibodies (Fig. 1D). Importantly, native AMPA receptor complexes in the cerebellum migrated at ∼669 kDa, which is similar to the size of GluA1 coexpressed with stargazin in oocytes (Vandenberghe et al., 2005). This result indicates that the AMPA receptor/stargazin complex is reconstituted in cRNA-injected oocytes on BN-PAGE.
During BN-PAGE, detergents bound to proteins, especially hydrophobic transmembrane proteins, have the effect of shifting protein migration to higher molecular weights. As such, transmembrane proteins often appear larger in molecular weight. In addition, unidentified interactions in a protein complex could render the molecular weight of a protein complex larger than expected. Therefore, it is not possible to deduce AMPA receptor stoichiometry from molecular-weight standards on BN-PAGE. Thus, we developed a novel strategy to determine the stoichiometry of the AMPA receptor and TARPs using BN-PAGE.
The AMPA receptor assembled as a tetramer that adopted a dimer-of-dimers conformation
Both GluA1 and GluA1ΔNTD functioned as glutamate-gated ion channels (Fig. 1B) and both structures were preserved on BN-PAGE as uniform complexes (Fig. 1D). The difference in the molecular weight of the two functional proteins on BN-PAGE was used to determine the stoichiometry of AMPA receptors. If two proteins assembled as hetero-oligomeric AMPA receptors without disrupting any other protein interactions, then the molecular weight of the resulting complex on BN-PAGE will be intermediate to the molecular weights of the two homo-oligomeric proteins. The number of subunits incorporated in each receptor complex was determined by counting the number of distinct molecular-weight bands between the homo-oligomers (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material).
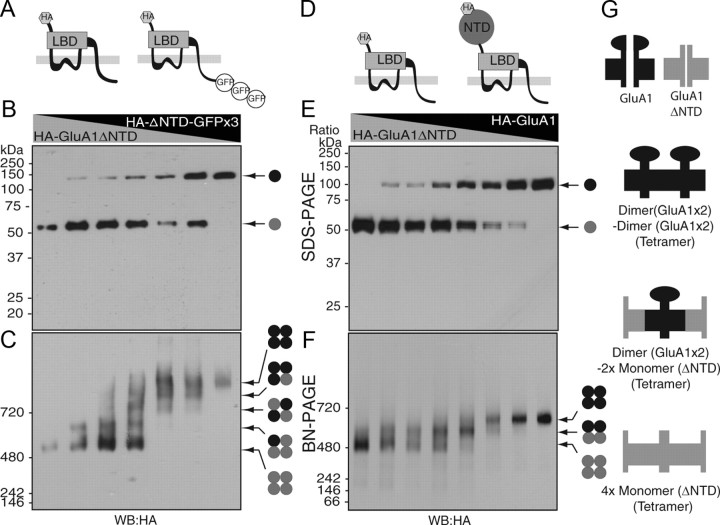
First, we used HA-GluA1ΔNTD and HA-GluA1ΔNTD fused to three monomeric GFP units (AcGFP×3; 90 kDa) since molecular weights of HA-GluA1ΔNTD and HA-GluA1ΔNTD-GFP×3 (50 kDa and 140 kDa, respectively) are significantly different without a disturbance in channel function (Fig. 2A). Xenopus laevis oocytes were injected with various ratios of HA-GluA1ΔNTD and HA-GluA1ΔNTD-GFP×3 cRNAs and then subjected to SDS-PAGE and BN-PAGE. GluA1ΔNTD and GluA1ΔNTD-GFP×3 were detected as single bands on SDS-PAGE in a cRNA dose-dependent manner (Fig. 2B). In contrast, five distinct bands were detected on BN-PAGE (Fig. 2C). This result led us to conclude that GluA1ΔNTD was a tetramer (Fig. 2C).
Figure 2.
The AMPA receptor formed a tetramer that adopted a dimer-of-dimers conformation. A, Schematic structure of GluA1ΔNTD tagged with HA epitope at the extracellular domain and with three monomeric units of AcGFP (GFP×3) at the C terminus. LBD, Ligand-binding domain. B, Oocytes were injected with various ratios of HA-GluA1ΔNTD and HA-GluA1ΔNTD-GFP×3 cRNAs [ratio of HA-GluA1ΔNTD (5 ng)/HA-GluA1ΔNTD-GFP×3 (13.3 ng), from the left: 1/0, 5/1, 3/1, 1/1, 1/5, 1/7, and 0/1]. HA-GluA1ΔNTD and HA-GluA1ΔNTD-GFP×3 were detected as single bands around 55 and 140 kDa, respectively, on SDS-PAGE. C, Five distinct bands were detected at a molecular weight >500 kDa (arrow) on BN-PAGE (4–10%). The presence of five distinct bands indicates that HA-GluA1ΔNTD and HA-GluA1ΔNTD-GFP×3 formed a tetramer. D, Schematic structure of GluA1 and GluA1ΔNTD, which were tagged with an HA epitope at the extracellular domain. E, Oocytes were injected with various ratios of HA-GluA1 and HA-GluA1ΔNTD cRNAs [ratio of HA-GluA1 (2 ng)/GluA1ΔNTD (2 ng), from the left: 1/0, 7/1, 3/1, 1/1, 1/3, 1/5, 1/7, and 0/1]. HA-GluA1 and HA-GluA1ΔNTD were detected as single bands around 100 and 55 kDa, respectively, on SDS-PAGE (10%). F, Three distinct bands were detected around 700, 600, and 500 kDa (arrow) on BN-PAGE (4–10%). The presence of three distinct bands indicates that GluA1 and GluA1ΔNTD formed a dimer. G, The NTD preferentially formed a dimer before tetramerization. The three distinct complexes of HA-GluA1 and HA-GluA1ΔNTD were a dimer of GluA1 dimers, a GluA1 dimer with two GluA1ΔNTD monomers, and four GluA1ΔNTD monomers. WB, Western blotting.
To determine the stoichiometry of full-length GluA1, we next injected various ratios of HA-GluA1 and HA-GluA1ΔNTD cRNAs into Xenopus laevis oocytes and performed SDS-PAGE and BN-PAGE (Fig. 2D). The expression of GluA1 and GluA1ΔNTD was confirmed on SDS-PAGE without any detectable protein degradation (Fig. 2E). Although HA-GluA1ΔNTD (∼480 kDa) was a tetramer (Fig. 2C), three (and not five) distinct bands of HA-GluA1 and HA-GluA1ΔNTD hetero-oligomers and homo-oligomers were detected using BN-PAGE (Fig. 2F). Similarly, anti-GluA1 antibody detected three distinct bands in oocytes injected with various combinations of GluA1 and GluA1ΔNTD (supplemental Fig. 2A–C, available at www.jneurosci.org as supplemental material). The difference in the molecular weight of each of the three distinct bands observed for HA-GluA1 and HA-GluA1ΔNTD hetero-oligomers was ∼90 kDa, which corresponds to two subunits of NTD (45 kDa). These results suggested that the NTD of full-length GluA1 preferentially forms a dimer before tetramerization. The three distinct complexes of HA-GluA1 and HA-GluA1ΔNTD (Fig. 2F) were a dimer of GluA1 dimers, a GluA1 dimer with two GluA1ΔNTD monomers, and four GluA1ΔNTD monomers (Fig. 2G).
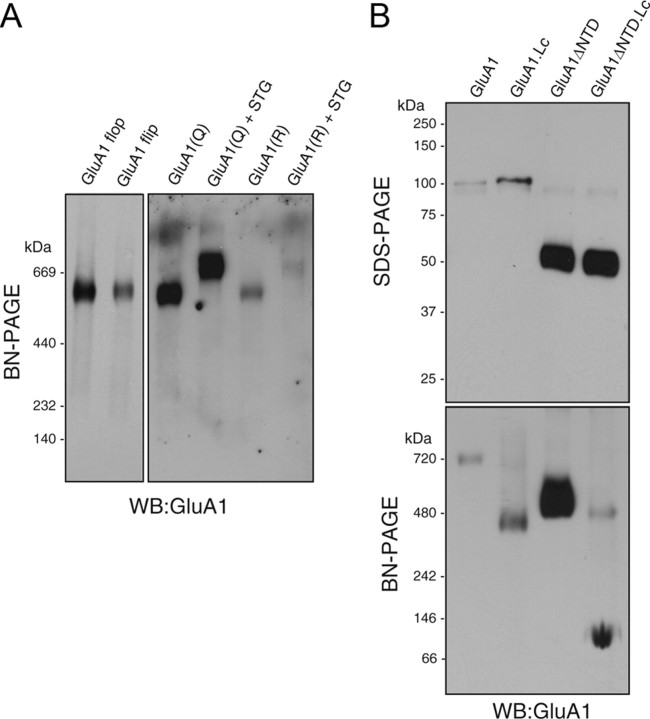
GluA1ΔNTD formed a tetramer from monomeric subunits instead of a dimer-of-dimers (Fig. 2C), which suggests that the NTD is the initial dimerization domain in the AMPA receptor. To identify a second dimerization domain in AMPA receptor dimers, we tested the effects of several AMPA receptor mutations on the assembly of the receptor. Neither flip/flop splicing variants located on the second extracellular loop of GluA1 nor mutations in the Q/R RNA editing site located in the pore loop affected the assembly of AMPA receptors (Fig. 3A). Interestingly, the GluA1 Lurcher mutant, which carries an A636T mutation near the second transmembrane domain, formed a tetramer less efficiently. Most of the GluA1 Lurcher mutants formed a dimer and most of the GluA1ΔNTD Lurcher mutants remained as monomers (Fig. 3B). This result suggests that the NTD dimerizes AMPA receptors as a first step and that sites around residue A636 of GluA1 are involved in the subsequent dimerization of two dimers. GluA1 formed a tetramer predominantly, whereas GluA1 with the Lurcher mutation and GluA1ΔNTD with the Lurcher mutation formed a dimer and a monomer, respectively (Fig. 3). These results suggest that GluA1 assembles predominantly as a tetramer, probably because GluA1 is predominantly tetrameric at steady state and not because GluA1 tetramers are more stable and monomers/dimers are degraded. Notably, similar to native AMPA receptors, we have detected a small proportion of dimers after long exposure (supplemental Fig. 2D, available at www.jneurosci.org as supplemental material), whereas AMPA receptors in transfected heterologous cells were detected predominantly as monomers and dimers (Penn et al., 2008). This difference is probably due to protein expression level.
Figure 3.
The Lurcher site of GluA1, but not the Q/R editing site or the flip/flop splicing isoforms, was involved in the second dimerization of AMPA receptor dimers. Oocytes were injected with the cRNAs indicated. A, Both GluA1 flip and flop splicing isoforms (5 ng of cRNA each) elicited single bands at around 669 kDa on BN-PAGE (4–10%). In addition, both GluA1 and the Q/R edited forms (5 ng of cRNA each) yielded single bands at around 669 kDa on BN-PAGE. Coinjection with stargazin (STG; 0.4 ng of cRNA) led to an increase in the molecular weight to a similar extent. B, GluA1 and GluA1ΔNTD carrying a mutation (A636T) corresponding to the Lurcher mutation in the Δ2 orphan receptor (GluA1 Lc or GluA1ΔNTD Lc, 5 ng of cRNA each) were observed as single bands at around 100 and 50 kDa, respectively, on 10% SDS-PAGE, which was similar to what was observed for the wild type. Although GluA1 Lc and GluA1ΔNTD Lc assembled as a tetramer, the majority of GluA1 Lc and GluA1ΔNTD Lc were detected on BN-PAGE (3–15%) as a dimer and a monomer, respectively. WB, Western blotting.
Variable stoichiometry of TARPs on AMPA receptors
Next, we explored the stoichiometry of TARPs on AMPA receptors. As stargazin is a relatively small protein (37 kDa) when compared with GluA1 (100 kDa), stargazin was fused with a large protein to allow adequate mobility shifts on PAGE. Therefore, we first examined stargazin tagged with a varying number of GFP units and confirmed the occurrence of molecular-weight shifts on BN-PAGE using oocytes coinjected with GluA1 cRNA. Despite the detection of a single band of GFP-tagged stargazin on SDS-PAGE, several distinct bands were detected as a GluA1 complex for stargazin tagged with multiple GFP units (data not shown). This result suggests that some GluA1 complexes contain a lesser number of stargazin units, which led us to speculate that the stargazin/GluA1 complex might exhibit variable stoichiometry.
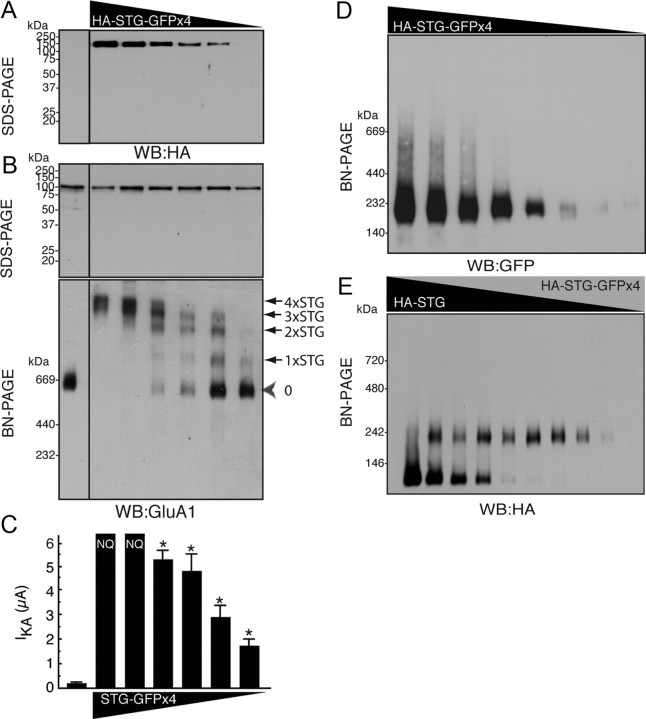
If the stoichiometry of stargazin on GluA1 is variable, we should detect a shift in the molecular weight of this protein complex that is dependent on the expression levels of stargazin (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). To examine this possibility, we expressed a fixed amount of GluA1 (2 ng) and varying amounts of stargazin tagged with an HA epitope in the first extracellular loop and with four monomeric GFP units in the cytoplasmic domain (HA-STG-GFP×4), the latter of which was expressed as a 150 kDa protein on SDS-PAGE (Fig. 4A). GluA1 was detected as a single band on SDS-PAGE, whereas four distinct bands were observed for the stargazin/GluA1 complex on BN-PAGE, depending on the expression levels of stargazin. We also detected stargazin-free AMPA receptors on BN-PAGE and noted that an increase in the expression levels of stargazin shifted GluA1/stargazin complexes to a higher molecular weight (Fig. 4B). Importantly, there seemed to be no cooperative interactions between stargazin and AMPA receptors, as the molecular weight of the stargazin complex increased linearly with the increase in the level of expression of stargazin (Fig. 4B). Furthermore, we measured AMPA receptor activity using TEVC recording to determine the number of stargazin units required for the modulation of AMPA receptor activity. We found that the concentration of stargazin (78 pg of cRNA) that led predominantly to a stoichiometry of one molecule of stargazin per AMPA receptor (1×STG) enhanced the kainate-evoked AMPA-receptor activity significantly compared with AMPA receptor alone (Fig. 4C). Lower stargazin concentrations (39 and 19 pg) also enhanced kainate-evoked AMPA receptor activity significantly (2 ng of GluA1 with no stargazin, 192 ± 63 nA; with 39 pg of stargazin, 645 ± 115 nA; with 19 pg of stargazin, 572 ± 94 nA). These results indicate that one stargazin molecule was sufficient to modulate the channel properties of AMPA receptors. In addition, HA-stargazin-GFP×4 did not form an oligomer upon high expression of HA-stargazin-GFP×4 (Fig. 4D). Furthermore, stargazin and stargazin-GFP×4 did not form heteromers on BN-PAGE, which suggests that stargazin was expressed as a monomer (Fig. 4E). These results imply that a maximum of four stargazin molecules bind to one AMPA receptor and that this is dependent on the expression levels of stargazin. Our results also indicated that one stargazin unit was sufficient to modulate the activity of the AMPA receptor.
Figure 4.
Variable stoichiometry of stargazin on AMPA receptors. A, Oocytes were injected with GluA1 cRNA (2 ng) and various amounts (from left to right: 0, 2.5, 1.25, 0.625, 0.313, 0.156, and 0.078 ng) of stargazin tagged with HA epitope at the extracellular domain and with four monomeric AcGFP units (GFP×4) at its cytoplasmic domain (HA-STG-GFP×4). Single bands of HA-stargazin-GFP×4 and GluA1 were detected on SDS-PAGE (10%) at around 150 kDa, as assessed using an anti-HA antibody, and at around 100 kDa, as assessed using an anti-GluA1 antibody. B, Up-regulation of the expression of HA-tagged stargazin (STG) containing four GFP units (HA-STG-GFP×4) shifted the molecular weight of the GluA1 complex detected using an anti-GluA1 antibody. We detected four distinct bands, in addition to a band corresponding to GluA1 alone, which suggests that a maximum of four stargazin molecules can bind to one AMPA receptor dimer-of-dimers structure. In addition, the molecular weight of the GluA1/stargazin complex was shifted linearly upon increase of stargazin expression, which suggests that all four stargazin-binding sites on the AMPA receptor were independent. C, Kainate-evoked (20 μm) currents from oocytes that were coinjected with GluA1 (2 ng) and various amounts of stargazin tagged with four GFP units (STG-GFP×4) were measured using TEVC recording. Data represent the means ± SEM (n = 5–7); asterisks indicate p < 0.05 using Tukey's test with ANOVA, a significant difference compared with GluA1 alone. NQ, non-quantitative because current amplitudes were too high (>12 μA). D, HA-stargazin tagged with four monomeric GFP units did not form oligomers in an expression-level-dependent manner (from left to right, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, and 0.031 ng) on BN-PAGE (4–15%). E, HA-tagged stargazin and HA-stargazin tagged with four monomeric GFP units did not form a dimer and were expressed as monomers on BN-PAGE (4–15%). Ratio of HA-STG (0.8 ng)/HA-STG-GFP×4 (1.6 ng), from left: 1/0, 7/1, 5/1, 3/1, 1/1, 1/3, 1/5, 1/7, and 0/1. WB, Western blotting.
Neuronal AMPA receptor complexes contained one TARP molecule
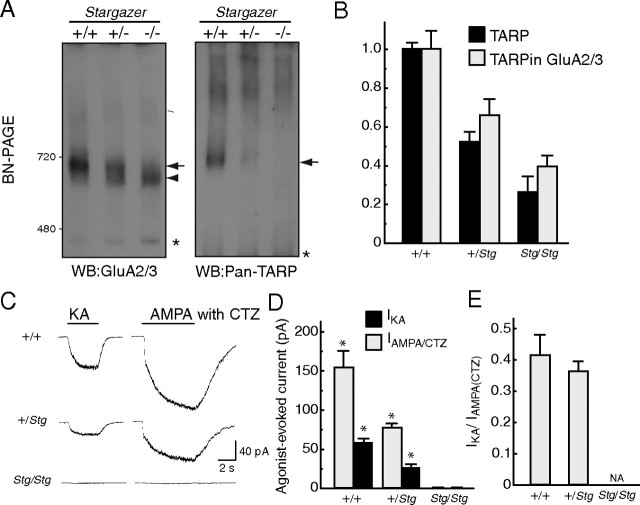
To determine the stoichiometry of stargazin on AMPA receptors in neurons, the simplest strategy would be to compare the molecular weights of neuronal and reconstituted AMPA receptor complexes. However, four AMPA receptor isoforms (GluA1–4) are expressed in the brain and their molecular weights differ from one isoform to the other, which complicates the interpretation of molecular-weight-based results. Therefore, we devised an alternative approach: we used stargazer mice, which have the advantage of lacking stargazin expression via an ETn insertion near exon 2 of the stargazin gene (Letts et al., 1998). Vandenberghe et al. (2005) showed that BN-PAGE can separate the AMPA receptor from the AMPA receptor associated with TARP (TARPin AMPA receptors) in the brain and found that the expression of TARPin AMPA receptor was decreased in the cerebellum in a stargazin copy-number-dependent manner. Importantly, quantitation of TARPs and TARPin AMPA receptors in different genotypes may reveal fixed or variable stoichiometry of TARPs in the brain in a TARP-expression-dependent manner. However, such systematic quantitation has not been performed. Therefore, we measured the ratio of TARPs and TARPin AMPA receptors in different stargazer genotypes to determine the fixed or variable nature of TARP stoichiometry on AMPA receptors.
The AMPA-receptor complex detected using the anti-GluA2/3 antibody exhibited two different sizes in neurons from wild-type and stargazer heterozygous mice (Fig. 5A), as shown previously (Vandenberghe et al., 2005). As there was no corresponding TARP signal detected using the anti-pan-TARP antibody (Fig. 5A), it is unlikely that the lower band detected in stargazer heterozygous neurons contained TARP. This result suggests that only the AMPA-receptor complex detected at the same size as the TARP complex was an AMPA receptor/TARP complex. Therefore, the higher band observed in heterozygous cells represented the TARPin AMPA receptor and the lower band corresponded to AMPA receptors without TARP (TARPless AMPA receptors) (Fig. 5A). In addition to these two bands detected on GluA2/3 Western blotting, we detected a band smaller than 480 kDa, as indicated by the asterisk, which could be a dimerized form of a GluA2/3.
Figure 5.
Neuronal AMPA receptors associated with one TARP/stargazin. Primary cerebellar granule cell cultures were prepared from littermates of wild-type (+/+), heterozygous (+/Stg), and homozygous (Stg/Stg) stargazer mice. A, Neurons were solubilized and resolved on 3–8% BN-PAGE. Two predominant bands of GluR2/3 in neurons from wild-type (left, arrow) and stargazer heterozygous (left, arrow and arrowhead) and homozygous (left, arrowhead) mice were observed on BN-PAGE. Stargazin was expressed in a copy-number-dependent manner (right, arrow). A single band of stargazin was detected in neurons from wild-type and heterozygous neurons, but not in neurons from homozygous stargazer mice. The asterisks indicate nonspecific bands, one of which could be GluA2/3 dimers. B, The signal intensity of GluA2/3 and stargazin bands in neurons from wild-type and stargazer heterozygous and homozygous mice was quantified using the ImageJ software and was normalized to the wild type. Importantly, the increase in the number of stargazin copies led to the concordant up-regulation of the AMPA receptor/TARP complex signal, which suggests a fixed stoichiometry of stargazin on AMPA receptors in neurons. C–E, Agonist-evoked whole-cell currents were measured in cultured mouse cerebellar granule cells. The steady-state currents evoked by 20 μm kainate (IKA) or 100 μm AMPA with 100 μm cyclothiazide (CTZ; IAMPA/CTZ) increased linearly in a stargazin copy-number-dependent manner. The representative traces (C), mean steady-state amplitude (D), and ratio of agonist-evoked currents (E) for each genotype are shown. Data represent the mean ± SEM (n = 4–5). Asterisks indicate p < 0.01 using Tukey's test with ANOVA, a significance difference of IKA or IAMPA/CTZ between wild-type and stargazer heterozygous or between stargazer heterozygous and homozygous. WB, Western blotting.
To examine AMPA receptor and TARP stoichiometry in neurons, we measured the signal intensity of TARP and TARPin AMPA receptors in three genotypic backgrounds (wild type, heterozygous, and homozygous) (Fig. 5B and supplemental Fig. 3, available at www.jneurosci.org as supplemental material). We found that the down-regulation of TARP in heterozygous stargazer cells correlated with reductions in the levels of TARPin AMPA receptors. The residual TARP complexes observed in homozygous stargazer cells could be due to the other TARP isoforms, γ-3 and γ-4, that were expressed in other types of cultured neurons (Fig. 5B). Importantly, the increase in stargazin copy number led to the concordant up-regulation of the AMPA receptor/TARP complex signal, which suggests a fixed stoichiometry of stargazin on AMPA receptors in neurons.
One of the important roles of TARP is the modulation of the channel properties of AMPA receptors. For example, TARP renders kainate more efficacious to AMPA receptors and increases the ratio of kainate- and glutamate-evoked currents (Tomita et al., 2005, 2007b; Turetsky et al., 2005; Korber et al., 2007). To this effect, we examined agonist-evoked currents. No agonist-evoked currents were detected in stargazer homozygous cerebellar granule cells. Kainate- and AMPA-evoked currents in neurons from wild-type mice were twice as large as those found in neurons of heterozygous mice, without changes in the ratio of kainate- and AMPA-evoked currents, which suggests that stargazin modulates AMPA-receptor activity in a stargazin copy-number-dependent manner (Fig. 5C,D). We did not observe any significant difference in the ratio of kainate- and AMPA with cyclothiazide-evoked currents between neurons from stargazer heterozygous and wild-type mice (Fig. 5E).
A fixed stoichiometry of TARP on neuronal AMPA receptors could be due to either saturating or minimal levels of TARP expression, i.e., one or four TARP molecules on one AMPA receptor. Importantly, we did not detect any unbound stargazin in wild-type and stargazer heterozygous mice, which suggests that neuronal stargazin expression levels do not allow a saturating association between AMPA receptors and the prototypical TARP, stargazin. Furthermore, we found no cooperative interaction between the four maximum stargazin units and the AMPA receptor (Fig. 4B), and one stargazin was sufficient to modulate AMPA-receptor activity (Fig. 4C). From these results, we concluded that only one stargazin interacts with one AMPA receptor tetramer, which forms a dimer-of-dimers structure, to modulate AMPA receptor activity in cerebellar granule cells.
Discussion
Here, we showed that functional AMPA receptors assembled as tetramers and formed a dimer-of-dimers structure biochemically. Using the same strategy, we also determined that the AMPA receptor auxiliary subunit, TARP, had a variable stoichiometry (1–4 TARP units) on AMPA receptors in a TARP amount-dependent manner. In cerebellar granule cells, only one TARP molecule interacted with the AMPA receptor and one TARP unit was sufficient to modulate AMPA-receptor activity. This fundamental composition of the AMPA receptor/TARP complex and the unique property of the TARP dose-dependent variable stoichiometry are important for the elucidation of the molecular machinery that underlies synaptic transmission.
Oligomerization of the AMPA receptor
Previous studies showed that the NTD of the AMPA receptor can dimerize and may contribute to the subunit-specific hetero-oligomerization of AMPA receptors (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001; Mansour et al., 2001; Jin et al., 2009; Kumar et al., 2009). In addition, the ligand-binding domain of AMPA receptors (also termed S1-S2 domain) can dimerize, especially after binding of cyclothiazide, which blocks the desensitization of AMPA receptors (Sun et al., 2002). Here, we found that the AMPA receptor formed a tetramer and adopted a dimer-of-dimers structure. In addition, we found that the NTD of the AMPA receptor mediated the first dimerizing interaction between AMPA receptor subunits. The second dimerization was mediated by domains located near the second transmembrane domain, which was supported by the less efficient tetramerization of the GluA1 Lurcher mutant (A636T). However, a small portion of GluA1 Lurcher mutant proteins formed tetramers, which suggests that A636T was involved in, but was not required for, the second dimerization. The A636T mutation may modulate the structure of the ligand-binding domain, which is considered one of the dimerization domains of the receptor.
We found that the first dimerization of the full-length AMPA receptor was mediated by its NTD. However, AMPA receptors lacking NTD retained channel activity (Pasternack et al., 2002; Tomita et al., 2007a). This indicates that the NTD was not necessary for AMPA receptor assembly. Most likely, the NTD plays roles in the subunit-specific assembly of AMPA receptors, as suggested previously (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001; Mansour et al., 2001). Moreover, the channel activity of GluA1ΔNTD suggests the presence of another dimerization/tetramerization domain in AMPA receptors, in addition to the NTD and ligand-binding domain. The identification of the domain that mediates the second dimerization of GluA1ΔNTD and of the full-length AMPA receptor is crucial and will require further investigation of the structure of the full-length AMPA receptor at the atomic level.
Variable stoichiometry of TARPs on AMPA receptors in heterologous cells
We found that TARPs adopt a variable stoichiometry (1–4 units) on AMPA receptors in heterologous systems in a TARP amount-dependent manner. Furthermore, each TARP molecule bound to AMPA receptors independently, without any cooperative binding properties, and one TARP unit was sufficient to modulate the activity of the AMPA receptor.
While we were finalizing this paper, another group published a similar study (Shi et al., 2009). These authors compared the ratios of kainate- and glutamate-evoked currents in AMPA receptor/TARP tandem proteins expressed in heterologous cells and concluded that AMPA receptors assume a variable stoichiometry and contain zero, two, or four units of TARP. This conclusion is consistent with our findings. In addition to two and four units of TARP on AMPA receptors, one and three units of TARP interacted with the AMPA receptor complex simultaneously (Fig. 4B). This odd number of TARP stoichiometry suggests that TARPs bind to AMPA receptor domains by preserving a fourfold symmetrical structure instead of a twofold symmetry. This result suggests that TARP may not be involved in either the first or the second dimerizations required for the formation of AMPA receptor tetramers.
Two isoforms of TARP homologous proteins, STG-1 and STG-2, were identified in Caenorhabditis elegans (Wang et al., 2008). Together with SOL-1, STG-1 and STG-2 modulate the channel activity of GLR-1 in cRNA-injected oocytes (Zheng et al., 2004, 2006; Walker et al., 2006a,b). However, coexpression of GLR-1 with either STG-1 or STG-2 led to different GLR-1 channel properties in cRNA-injected oocytes. This result suggests that GLR-1 assembles with more than two TARPs and is consistent with our result showing that one AMPA receptor can associate with more than two TARPs, depending on the levels of expression of TARP. It is important to elucidate how many TARP-like STG units are incorporated into the GLR-1 complex in vivo.
Stoichiometry of TARPs on AMPA receptors in neurons
In cerebellar granule cells, we found that TARP had a fixed and minimum stoichiometry on AMPA receptors. Because the minimum number of TARP units necessary to modulate AMPA receptor activity is one, it is very likely that neuronal AMPA receptors contain only one TARP per AMPA receptor in cerebellar granule cells.
Independently, a recent paper by Shi et al. (2009) showed that neuronal AMPA receptors take on a variable stoichiometry and contain zero, two, or four TARP units, by comparing the ratios of kainate- and glutamate-evoked currents in AMPA receptor/TARP tandem proteins expressed in heterologous cells, as well as in neuronal AMPA receptors. The disparity between their conclusions and ours (i.e., fixed and variable stoichiometries of TARPs) could be due to the neuronal type studied; we used cerebellar cells, while Shi et al. used hippocampal cells. We did not detect a cooperative interaction between TARPs and the AMPA receptor (Fig. 4B). This indicates that the number of TARP units on the AMPA receptor was dependent on the expression levels of TARP and that the stoichiometry of TARPs on AMPA receptors could vary according to brain region. The systematic quantitative analysis of TARPs and AMPA receptors will be required to elucidate the detailed mechanisms that underlie this process.
One important role of TARPs is to modulate AMPA-receptor activity. Here, we found that one TARP was sufficient to modulate AMPA-receptor activity, including the ratio of kainate- and glutamate-evoked currents. However, this ratio of agonist-evoked currents varies considerably between the AMPA receptor splicing isoforms, flip and flop, which affects the ratios of kainate- and glutamate-evoked currents significantly (Jonas and Sakmann, 1992; Partin et al., 1995; Koike et al., 2000; Vorobjev et al., 2000; Tomita et al., 2005). A characterization of the channel properties of flop splicing isoforms of AMPA receptors would enable a comparison of agonist-evoked currents among neurons.
A previous study used coimmunoprecipitation experiments to demonstrate that each of the four class I TARPs (stargazin/γ-2, γ-3, γ-4, and γ-8) was not included in the same AMPA-receptor complex in the cerebellum (Tomita et al., 2003). There are three possible explanations for this phenomenon: (1) differential expression of each TARP in different neurons of the cerebellum, (2) preferential assembly of a single TARP isoform in one AMPA receptor complex, and (3) presence of only one TARP in a single AMPA-receptor complex. Although each TARP isoform is expressed in distinct neurons of the cerebellum (Tomita et al., 2003; Fukaya et al., 2005), some neurons, including Purkinje cells, express more than two TARP isoforms and heteromeric TARP complexes should be detectable. Therefore, TARPs may form homomeric TARP complexes preferentially, via the AMPA receptor, or there may be one TARP in the AMPA receptor complex in the cerebellum.
Stoichiometry of TARPs on AMPA receptors at synapses
The amplitude and decay of AMPA receptor-mediated miniature EPSCs is slightly, but significantly, different in cerebellar granule neurons from wild-type and stargazer heterozygous mice (Milstein et al., 2007). This could be caused by differences in the stoichiometry of stargazin on AMPA receptors at synapses or by the presence of different populations of TARPin and TARPless AMPA receptors at synapses.
TARP/stargazin is required for surface expression of AMPA receptors in cerebellar granule cells (Hashimoto et al., 1999; Chen et al., 2000). However, glutamate-induced desensitization of AMPA receptors causes decoupling of TARPs from functional AMPA receptors (Tomita et al., 2004; Morimoto-Tomita et al., 2009), i.e., there are two populations of AMPA receptors, TARPin and TARPless AMPA, at the cell surface.
Alternatively, a small proportion of AMPA receptors in wild-type neurons may contain more than one TARP and AMPA receptors containing more TARPs traffic better to synapses. The detection of the number of TARPs on one AMPA receptor at synapses is necessary to address this possibility.
Recently, several proteins were identified as subunits of ionotropic glutamate receptors (Jackson and Nicoll, 2009; Tigaret and Choquet, 2009). For example, cornichon on AMPA receptors, NETO1 and NETO2 as kainate receptor regulatory proteins on kainate receptors, and NETO1 on NMDA receptors (Ng et al., 2009; Schwenk et al., 2009; Zhang et al., 2009). It will be important to elucidate the differences in the assembly and stoichiometry of the subunits of ionotropic glutamate receptors identified recently.
Footnotes
S.T. is supported by the National Institutes of Health Grant MH07793, an Alfred P. Sloan research fellowship, a National Alliance for Research on Schizophrenia and Depression Young Investigator Award, the Esther A. and Joseph Klingenstein Fund, and the Edward Mallinckrodt Jr. Foundation. We thank Dr. Jim Howe and members of the Tomita laboratory for critical discussion.
References
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Coombs ID, Cull-Candy SG. Transmembrane AMPA receptor regulatory proteins and AMPA receptor function in the cerebellum. Neuroscience. 2009;162:656–665. doi: 10.1016/j.neuroscience.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Yamazaki M, Sakimura K, Watanabe M. Spatial diversity in gene expression for VDCCgamma subunit family in developing and adult mouse brains. Neurosci Res. 2005;53:376–383. doi: 10.1016/j.neures.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron. 2002;34:759–772. doi: 10.1016/s0896-6273(02)00693-1. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB. Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron. 2006;51:85–97. doi: 10.1016/j.neuron.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Fukaya M, Qiao X, Sakimura K, Watanabe M, Kano M. Impairment of AMPA receptor function in cerebellar granule cells of ataxic mutant mouse stargazer. J Neurosci. 1999;19:6027–6036. doi: 10.1523/JNEUROSCI.19-14-06027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. Neuroscience: AMPA receptors get “pickled.”. Nature. 2009;458:585–586. doi: 10.1038/458585a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Sakmann B. Glutamate receptor channels in isolated patches from CA1 and CA3 pyramidal cells of rat hippocampal slices. J Physiol. 1992;455:143–171. doi: 10.1113/jphysiol.1992.sp019294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S. Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci. 2000;20:2166–2174. doi: 10.1523/JNEUROSCI.20-06-02166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber C, Werner M, Kott S, Ma ZL, Hollmann M. The transmembrane AMPA receptor regulatory protein γ4 is a more effective modulator of AMPA receptor function than stargazin (γ2) J Neurosci. 2007;27:8442–8447. doi: 10.1523/JNEUROSCI.0424-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett FS, 2nd, Mori Y, Campbell KP, Frankel WN. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem. 1999;274:16907–16916. doi: 10.1074/jbc.274.24.16907. [DOI] [PubMed] [Google Scholar]
- Mano I, Teichberg VI. A tetrameric subunit stoichiometry for a glutamate receptor-channel complex. Neuroreport. 1998;9:327–331. doi: 10.1097/00001756-199801260-00027. [DOI] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Kamiya Y, Yuzaki M. Roles of the N-terminal domain on the function and quaternary structure of the ionotropic glutamate receptor. J Biol Chem. 2005;280:20021–20029. doi: 10.1074/jbc.M410513200. [DOI] [PubMed] [Google Scholar]
- Midgett CR, Madden DR. The quaternary structure of a calcium-permeable AMPA receptor: conservation of shape and symmetry across functionally distinct subunit assemblies. J Mol Biol. 2008;382:578–584. doi: 10.1016/j.jmb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Milstein AD, Zhou W, Karimzadegan S, Bredt DS, Nicoll RA. TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron. 2007;55:905–918. doi: 10.1016/j.neuron.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Zhang W, Straub C, Cho CH, Kim KS, Howe JR, Tomita S. Autoinactivation of neuronal AMPA receptors via glutamate-regulated TARP interaction. Neuron. 2009;61:101–112. doi: 10.1016/j.neuron.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Ng D, Pitcher GM, Szilard RK, Sertié A, Kanisek M, Clapcote SJ, Lipina T, Kalia LV, Joo D, McKerlie C, Cortez M, Roder JC, Salter MW, McInnes RR. Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol. 2009;7:e41. doi: 10.1371/journal.pbio.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- Osten P, Stern-Bach Y. Learning from stargazin: the mouse, the phenotype and the unexpected. Curr Opin Neurobiol. 2006;16:275–280. doi: 10.1016/j.conb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Partin KM, Bowie D, Mayer ML. Structural determinants of allosteric regulation in alternatively spliced AMPA receptors. Neuron. 1995;14:833–843. doi: 10.1016/0896-6273(95)90227-9. [DOI] [PubMed] [Google Scholar]
- Pasternack A, Coleman SK, Jouppila A, Mottershead DG, Lindfors M, Pasternack M, Keinanen K. Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J Biol Chem. 2002;277:49662–49667. doi: 10.1074/jbc.M208349200. [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Irizarry SN, Hughes TE, Howe JR. Subunit interactions and AMPA receptor desensitization. J Neurosci. 2001;21:5574–5586. doi: 10.1523/JNEUROSCI.21-15-05574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Safferling M, Tichelaar W, Kummerle G, Jouppila A, Kuusinen A, Keinanen K, Madden DR. First images of a glutamate receptor ion channel: oligomeric state and molecular dimensions of GluRB homomers. Biochemistry. 2001;40:13948–13953. doi: 10.1021/bi011143g. [DOI] [PubMed] [Google Scholar]
- Sager C, Tapken D, Kott S, Hollmann M. Functional modulation of AMPA receptors by transmembrane AMPA receptor regulatory proteins. Neuroscience. 2009;158:45–54. doi: 10.1016/j.neuroscience.2007.12.046. [DOI] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klöcker N. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Seeburg PH. The TINS/TiPS Lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lu W, Milstein AD, Nicoll RA. The stoichiometry of AMPA receptors and TARPs varies by neuronal cell type. Neuron. 2009;62:633–640. doi: 10.1016/j.neuron.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Tigaret C, Choquet D. Neuroscience. More AMPAR garnish. Science. 2009;323:1295–1296. doi: 10.1126/science.1171519. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, Bredt DS. Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Tomita S, Shenoy A, Fukata Y, Nicoll RA, Bredt DS. Stargazin interacts functionally with the AMPA receptor glutamate-binding module. Neuropharmacology. 2007a;52:87–91. doi: 10.1016/j.neuropharm.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Tomita S, Byrd RK, Rouach N, Bellone C, Venegas A, O'Brien JL, Kim KS, Olsen O, Nicoll RA, Bredt DS. AMPA receptors and stargazin-like transmembrane AMPA receptor-regulatory proteins mediate hippocampal kainate neurotoxicity. Proc Natl Acad Sci U S A. 2007b;104:18784–18788. doi: 10.1073/pnas.0708970104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetsky D, Garringer E, Patneau DK. Stargazin modulates native AMPA receptor functional properties by two distinct mechanisms. J Neurosci. 2005;25:7438–7448. doi: 10.1523/JNEUROSCI.1108-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA, Bredt DS. Stargazin is an AMPA receptor auxiliary subunit. Proc Natl Acad Sci U S A. 2005;102:485–490. doi: 10.1073/pnas.0408269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev VS, Sharonova IN, Haas HL, Sergeeva OA. Differential modulation of AMPA receptors by cyclothiazide in two types of striatal neurons. Eur J Neurosci. 2000;12:2871–2880. doi: 10.1046/j.1460-9568.2000.00175.x. [DOI] [PubMed] [Google Scholar]
- Walker CS, Francis MM, Brockie PJ, Madsen DM, Zheng Y, Maricq AV. Conserved SOL-1 proteins regulate ionotropic glutamate receptor desensitization. Proc Natl Acad Sci U S A. 2006a;103:10787–10792. doi: 10.1073/pnas.0604520103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CS, Brockie PJ, Madsen DM, Francis MM, Zheng Y, Koduri S, Mellem JE, Strutz-Seebohm N, Maricq AV. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci U S A. 2006b;103:10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Walker CS, Brockie PJ, Francis MM, Mellem JE, Madsen DM, Maricq AV. Evolutionary conserved role for TARPs in the gating of glutamate receptors and tuning of synaptic function. Neuron. 2008;59:997–1008. doi: 10.1016/j.neuron.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Liu CI, Chang YC. A study of the oligomeric state of the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-preferring glutamate receptors in the synaptic junctions of porcine brain. Biochem J. 1996;319:731–739. doi: 10.1042/bj3190731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, Tomita S. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV. SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature. 2004;427:451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Walker CS, Francis MM, Maricq AV. SOL-1 is an auxiliary subunit that modulates the gating of GLR-1 glutamate receptors in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103:1100–1105. doi: 10.1073/pnas.0504612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53:627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]