Figure 1.
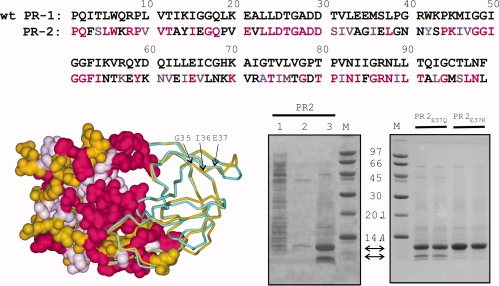
(Top) Sequence homology between wild-type PR1 (without stabilizing mutations) and PR2 (PDB code: 3EBZ) showing identical residues (magenta) and conservative substitutions (light purple). (Bottom left) Structural overlay of PR2 (gold) with autoproteolysis-resistant PR1 (PDB code: 2IEN, shown in cyan). Both crystal structures contain the inhibitor DRV (not shown) in the active site. The left half of the structure shows a surface representation of PR2 with identical residues and conservative substitutions relative to PR1-colored magenta and light purple, respectively. The major site of autoproteolysis, G35/I36, and residue E37 that was mutated to restrict autoproteolysis are indicated by arrows. (Bottom right) Expression of PR2 and its mutants in E. coli and analyses by SDS-PAGE. Lane 1, total soluble extract after initial lysis and centrifugal fractionation. The insoluble pellet obtained from the prior step was briefly sonicated in buffer containing 1M urea and 0.5% NP-40 (see “Materials and Methods” for complete composition) and the supernatant (lane 2) and insoluble (inclusion bodies, lane 3, and duplicate lanes of proteins derived from PR2E37Q and PR2E37K mutants) fractions derived after centrifugation are shown. Proteins were subjected to 20% homogeneous PhastGel electrophoresis followed by coomassie staining.
