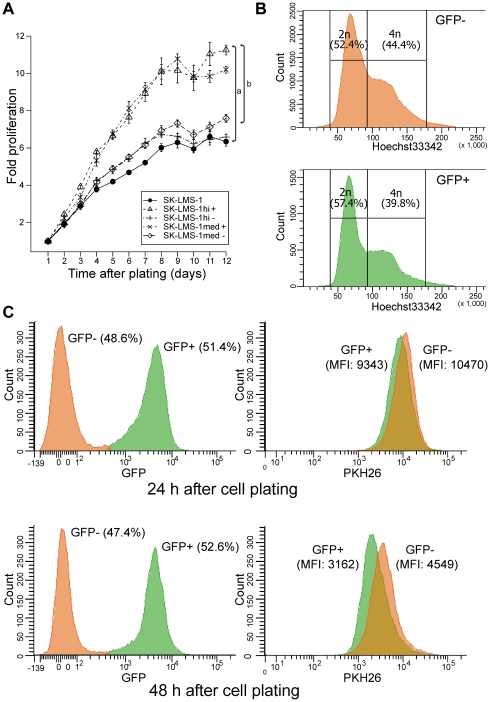
Figure 12. Knockdown of Kaiso in SK-LMS-1 cells accelerates cell proliferation.
(A) SK-LMS-1 parental cells and various SK-LMS-1_shKAISO derivative cell populations were subjected to an SRB cell proliferation assay. Cells with medium to high GFP expression levels (med+ and hi+; with stringent Kaiso knockdown) did proliferate more rapidly and reached significantly higher saturation densities than cells with very low GFP expression (hi– and med–; with poor Kaiso knockdown). Values are means ± SD; n = 6. Letters a and b indicate significant differences (p<0.05; Wilcoxon rank sum test): a, SK-LMS-1hi+ vs. parental cells; b, SK-LMS-1hi+ vs. SK-LMS-1hi- cells. (B) Cell cycle profile from unsynchronized SK-LMS-1_shKAISO cell populations with either low GFP expression (GFP–; with poor Kaiso knockdown) or high GFP expression (GFP+, with stringent Kaiso knockdown). Cells were stained with Hoechst33342 and analyzed by flow cytometry to detect 2n/4n DNA contents. In the GFP+ cells, fewer cells (39.8%) contained 4n amounts of DNA than in the GFP– cells (44.4%). Data shown represent one example of four independent experiments. All analyses were performed on subconfluent cell populations. (C) Flow cytometric analysis of cell proliferation by unsynchronized SK-LMS-1_shKAISO cell populations. At time 0, cells were stained with the red fluorescent cell-tracking dye, PKH26, which incorporates into cell membrane lipids and is thus diluted twofold every cell division. At 24 h and at 48 h after plating, cells were first distinguished according to GFP levels (left panels), where cells with high GFP levels (GFP+) show strong Kaiso knockdown. Dead cells were excluded from analysis by positive staining for SytoxRed. Mean PKH26 fluorescence intensity (MFI) was then measured in each subpopulation (right panels), showing that GFP+ cells underwent 1.56 [log2(9343/3162)] cell divisions in 24 h, whereas GFP– cells underwent 1.20 [log2(10470/4549)] cell divisions.