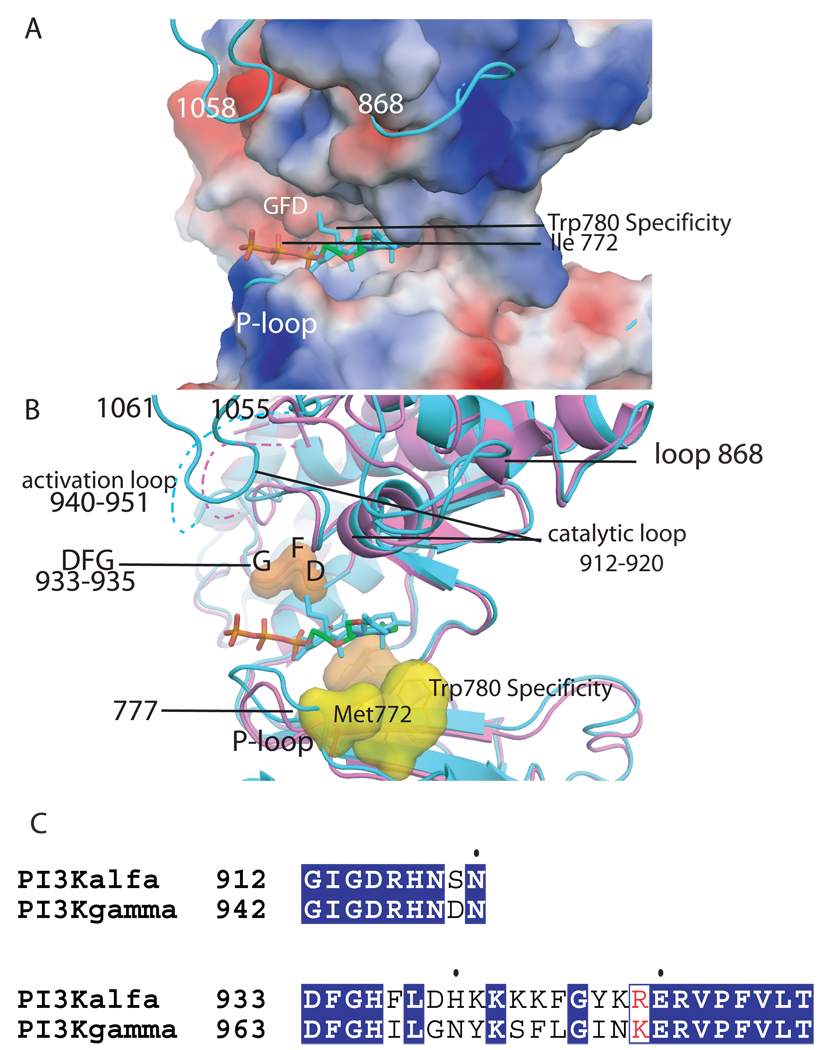
Figure 2. ATP binding site of the kinase domain of p110α.
A. Electrostatic surface of the kinase domain as in the wild type p110α/niSH2 (pdb id 2RD0) structure with the ATP modeled based on the p110γ complex structure (pdb id 1E8X). The complex of wortmannin bound H1047R p110α/niSH2 is shown in turquoise to highlight the conformational changes in loops 772–777 (P-loop), 864–872 and 1047–1062 which protrude from the surface. B. Structural overlap of the wild type p110α/niSH2 (magenta, pdb id 2RD0) structure with the wortmannin bound H1047R p110α/niSH2 (turquoise, 3HMM) overlapping a molecule of ATP. The recognition and catalytically important residues are shown as surfaces. The DFG (residues 933–935) is shown in orange; the protein kinase “gatekeeper residue” in p110γ (Ile 848) is shown in light blue; specificity residues Met 772 and Trp780 are shown in yellow; the activation loop is shown in both structures as a dashed line; the last residues of the p110α H1047R mutant which change conformations in the mutant are also shown in turquoise. C. Sequence alignment of the activation and catalytic loop of p110α and p110γ with identical residues shown in blue background.