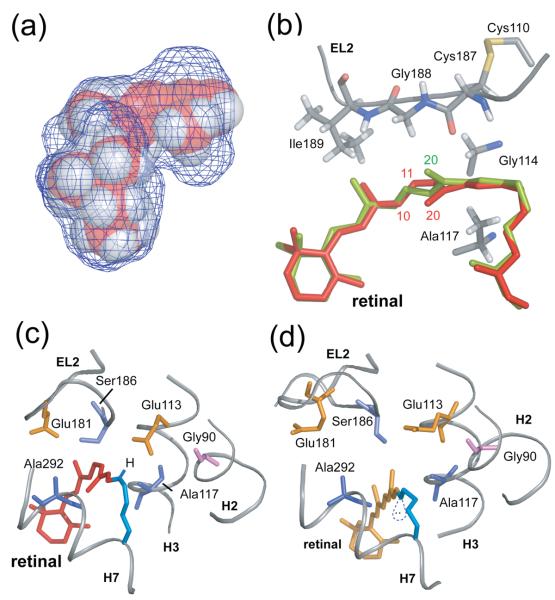
Fig. 2.
Trajectory of the C20 methyl group and Schiff base proton upon retinal isomerization. (A) The packing of residues around the retinal chromophore in the crystal structure of rhodopsin is visualized by calculating a cavity surface for the binding site. The shape of this cavity likely determines the motion of the C19 and C20 methyl groups upon isomerization. The C19 methyl group is tightly packed against Ile189 and Tyr191, whereas a void exists above the retinal C20 methyl group in the direction of the EL2 loop. The view in this panel is from the β-ionone ring end of the retinal. (B) Structure of the retinal binding site in rhodopsin highlighting the position of the retinal in rhodopsin (red) and in a model from MD simulations immediately after isomerization (orange). (C) and (D) present views of the retinal binding site in rhodopsin viewed from the Schiff base end of the retinal highlighting the position, orientation and interactions of the retinal SB nitrogen. (C) In rhodopsin, the SB proton is oriented toward the extracellular side of the receptor and interacts with Glu113 (H3) and Ser186 (EL2). (D) In the guided MD simulations of Meta II, the electron pair on the SB nitrogen has rotated to a more hydrophobic environment and is pointing toward the cytoplasmic side of the protein.