Fig. 8.
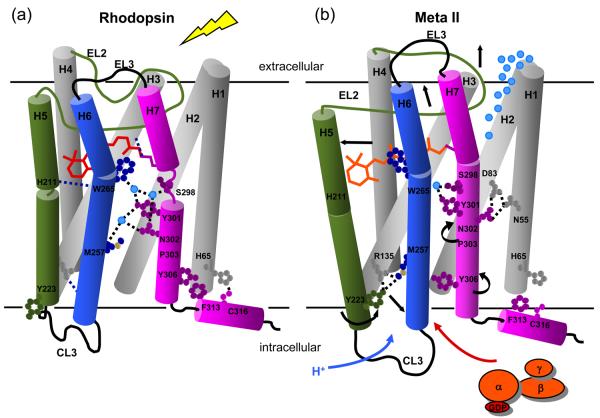
Working model of rhodopsin activation. Retinal isomerization leads to the strain in retinal binding site; all-trans retinal is not accommodated in the binding site in the dark. There are several studies probing the sequence of structural changes following isomerization. In detergent, Hofmann and coworkers118,119 have shown that SB deprotonation precedes the motion of H6 and proton uptake by Glu134. However, changes in the IR bands of Glu122 already accompany the formation of Meta I120 and the retinal SB in Meta I is accessible to hydroxylamine.121 Together these observations suggest that retinal isomerization first leads to displacement of EL2 and (at least limited) motion of H5 prior to deprotonation. Deprotonation of the SB then allows the retinal to shift into an active position that either guides or drives rotation of Trp265 and H6. This sequence of events is consistent with kinetic FTIR measurements122 indicating that there may be 15-20% deprotonation of the retinal Schiff base before changes in the IR bands between 1755 and 1740 cm−1 associated with Asp83 and Glu122. Rotation of Trp265 side chain towards EL2 disrupts the network of hydrogen bonding interactions involving the NPxxY motif on H7 and responsible for maintaining the inactive state of the receptor. The outward rotation of H6 is accompanied by the rotation of Tyr223 and Tyr306 into the H3-H6 interface.12 These residues face Met257 and hydrogen bond with Arg135, disrupting the ionic lock.