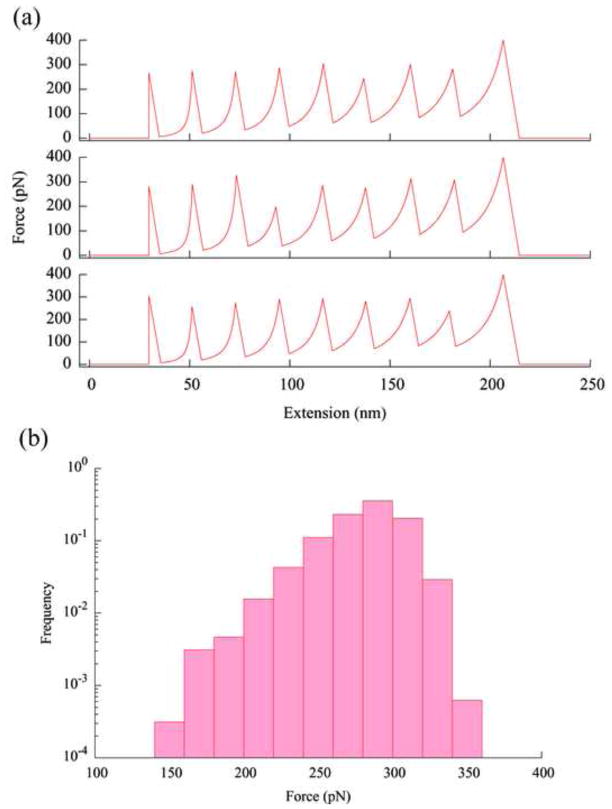
Fig. 2.
(a) Three simulated force curves from pulling a polymer of eight identical protein molecules. Simulation was carried out using these parameters: pulling speed v = 1 μm/s, cantilever spring constant κ = 50 pN/nm, temperature T = 300 K, persistence length of unfolded proteins pu = 0.40 nm, Δxu = 0.225 nm, and ku0 = 5×10−5 s−1. The contour length between the two linking point is Lu=28.1 nm in an unfolded protein and the distance between these two points in a folded protein is Lf =3.7 nm. These parameters are those used in the experiments of pulling ubiquitin molecules connected through the N-C termini [10,11]. Detachment from the tip is assumed to occur at a force of 400 pN. In experiments, detachments have been observed to occur at a variety of forces. (b) The distribution of the unfolding forces from 400 simulated force curves (3200 data points) as those shown in (a). The frequency is normalized by the total number of points, i.e., the height of each bin is equal to the number of data points in that bin divided by the total number of data points (3200, for this histogram).