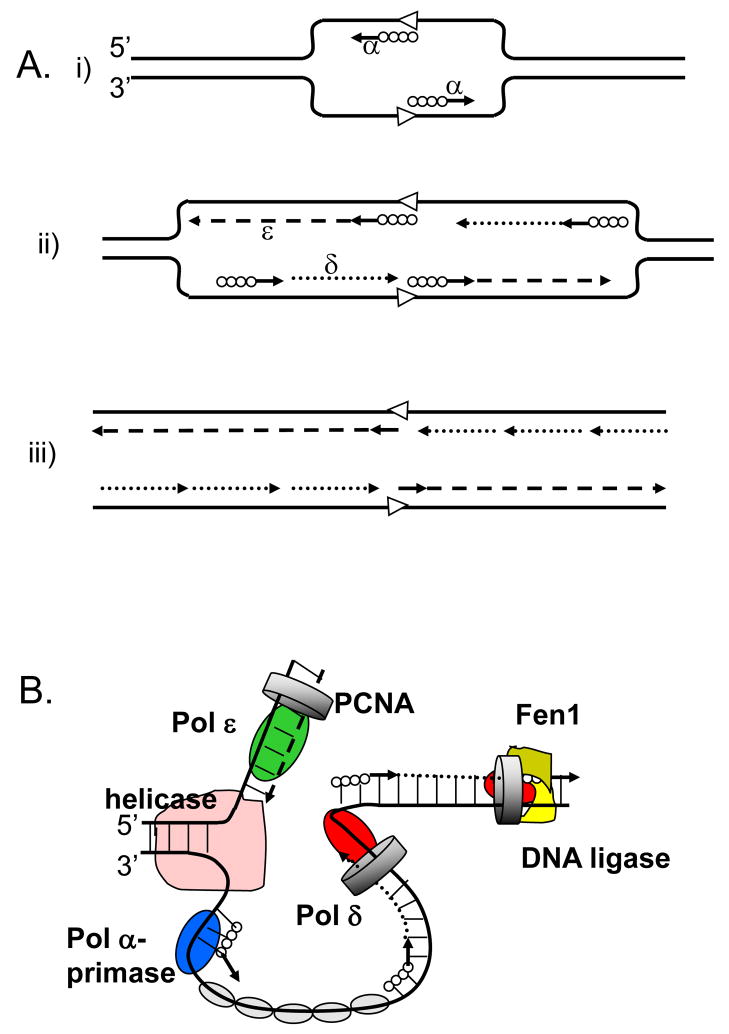
Figure 1. Three-polymerase model of replication fork in eukaryotes.
A. The model proposed in 1990 by Sugino group (adapted from [2].
(i) Pol α synthesizes short DNA segments (straight lines) primed by RNA (solid circles) at the replication origin (open arrowheads). (ii) Pol ε synthesizes the leading strand (dashed line); Pol α synthesizes short RNA-DNA stretches on the lagging strand, which are subsequently extended by Pol δ. (iii) After removal of RNA primers, Pol δ completes the lagging strand synthesis (dotted lines).
B. The currently accepted model (adapted from [85]).
The model illustrates primary roles for Pol ε (green oval) in leading DNA strand replication (dashed line) and Pol δ (red oval) in lagging strand replication (dotted line). Other proteins shown include the Pol α-primase (blue oval) synthesizing RNA-DNA hybrids (solid circles and straight lines), the MCM helicase (pink), the eukaryotic single-stranded-DNA-binding protein, replication protein A (RPA; gray ovals), the sliding clamp proliferating cell nuclear antigen (PCNA; gray ring) and the Fen1–DNA ligase complex (khaki-yellow).