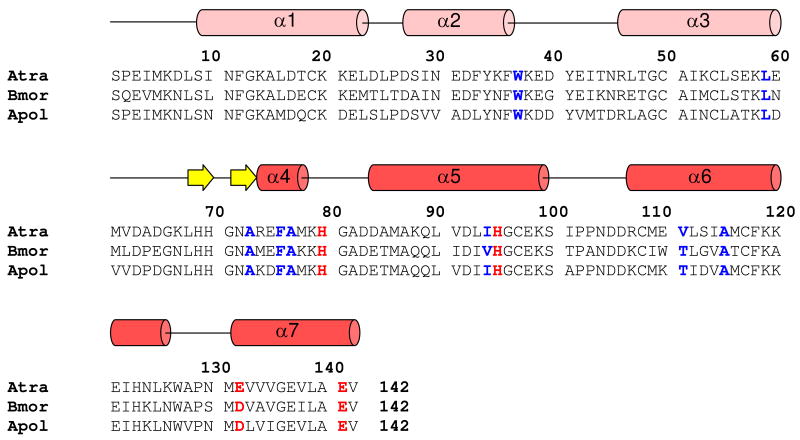
Figure 1.
Amino acid sequence alignment of AtraPBP1 with other moth PBPs. Residues forming histidine protonation switch are highlighted in bold and red. Hydrophobic residues interacting with α7 and implicated in pheromone binding are colored blue. Secondary structural elements at pH 4.5 indicated schematically (helices in red and light-red; strands in yellow) were derived from the analysis of NMR data (3JHNHα, chemical shift index, and sequential NOE patterns).