Abstract
The higher fungi exhibit a dichotomy with regard to urea utilization. The hemiascomycetes use urea amidolyase (DUR1,2) whereas all other higher fungi use the nickel-containing urease. Urea amidolyase is an energy dependent biotin-containing enzyme. It likely arose prior to the Euascomycete/Hemiascomycete divergence ca. 350 million years ago by insertion of an unknown gene into one copy of a duplicated methylcrotonyl CoA carboxylase (MccA). The dichotomy between urease and urea amidolyase coincides precisely with that for the Ni/Co transporter (Nic1p) which is present in the higher fungi that use urease and absent in those that do not. We suggest that the selective advantage for urea amidolyase is that it allowed the hemiascomycetes to jettison all Ni2+ and Co2+ dependent metabolism and thus to have two fewer transition metals whose concentrations need to be regulated. Also, the absence of MccA in the hemiascomycetes coincides with and may explain their production of fusel alcohols.
Introduction
We have long been interested in the role of nitrogen sources in regulating fungal dimorphism. For instance, the growth morphology of Ceratocystis ulmi and Trigonopsis variabilis could be modulated by the source of nitrogen. For C. ulmi, the cells grew as yeasts with proline and as hyphae with ammonia, arginine, and most other nitrogen sources (Kulkarni and Nickerson, 1981) while for T. variabilis, the cells grew as budding yeasts with ammonium sulfate and as triangles with methionine (Sentheshanmuganathan and Nickerson, 1962). One nitrogen source that has been understudied in C. albicans is urea. This inattention likely derives from numerous reports that C. albicans lacks urease (Odds, 1988) even though Dastidar et al (1967) reported that most strains of C. albicans grew well with urea as the sole source of nitrogen. A partial resolution of this impasse derives from the fact that C. albicans uses urea amidolyase to hydrolyze urea (Ghosh et al, 2009). The enzyme urea amidolyase, encoded by DUR 1, 2 (Degradation of URea), was first characterized in the yeast Candida utilis (Roon and Levenberg, 1972). This cytoplasmic, biotin-dependent enzyme (Roon et al, 1972) consists of a single protein chain with domains for both urea carboxylase and allophanate hydrolase activity (Cooper et al, 1980).
urea → 2NH3 + CO2 urease
urea + ATP + HCO3− → allophanate + ADP + Pi urea carboxylase
allophanate → 2NH3 + 2CO2 allophanate hydrolase
In Saccharomyces cerevisiae the actual inducer for DUR1,2 is allophanate, also known as urea carboxylate, rather than urea itself (Cooper, 1982). Allophanate is the chemical intermediate for the multifunctional Dur1, 2p, see equations (ii) and (iii). Here we report the conservation of urea utilization in fungi and the phylogenetic distribution of urease and urea amidolyase in the fungi.
Distribution of Urease and Urea Amidolyase in the Fungi
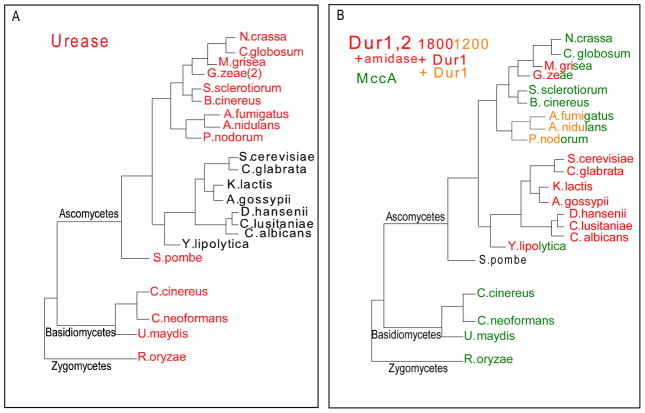
To identify which fungi have urease and which have urea amidolyase, we examined 22 available fungal genomes spanning the Ascomycetes and Basidiomycetes for the presence of the respective genes. There is a dichotomy. Urease (Fig. 1A in red) was found in all of the fungi except for members of the hemiascomycetes (Fig. 1A in black). The hemiascomycetes are those Ascomycetes which do not form fruiting bodies. The results are consistent with loss of the urease gene sometime prior to the Euascomycete – Hemiascomycete divergence ca. 350 million years ago (Galagan et al, 2005).
Fig. 1.
Phylogeny of fungal urease, urea amidolyase, (Dur1,2), and methylcrotonyl-CoA carboxylase. Sequences of fungal proteins were obtained from NCBI (www.ncbi.nlm.nih.gov) and the Fungal Genome Initiative (www.broad.mit.edu/annotation/fgi/). All BLAST searches were conducted using default parameters. MacVector software (Oxford Molecular Sciences, Inc., Hunt Valley, MD) was used for processing and analysis of sequences. The dendogram was prepared in PowerPoint and represents the current view of fungal phylogeny as presented by James et al. (2006). 1A. The presence (red) or absence (black) of urease homologues was identified using Cryptococcus neoformans URE1 (AF006062) as a query for BLASTp searches. 1B. The presence of full-length Dur1,2 (i.e., ~1800 amino acids) is indicated in red, intermediate length Dur1,2 (i.e., ~1200 amino acids) is indicated in orange, and methylcrotonyl-CoA carboxylase is indicated in green. Split red-green and orange-green labels indicate species that contain both Dur1,2 and methylcrotonyl-CoA carboxylase while black indicates that none of the above genes was detected. Dur1, 2 and MccA homologues were identified using S. cerevisiae Dur 1, 2 (CAA85172) as a query for BLASTp searches.
The dual function urea amidolyase Dur 1, 2 comes in two sizes: the longer of ca. 1800 aa (Fig. 1B in red) has an ca. 600 aa “amidase” domain fused at the N-terminus, whereas the shorter of ca. 1200 aa (Fig. 1B in orange) has not. The longer DUR 1, 2 is present in all hemiascomycetes examined, including C. albicans, whereas both the longer and shorter versions are found in subsets of the euascomycetes (Fig. 1B). One explanation is that DUR 1, 2 arose via duplication of the gene for MccA, mitochondrial biotin-containing methylcrotonyl-CoA carboxylase (ca. 700 aa), followed by fusion of one of the MccA genes with another still unidentified gene (ca. 500 aa) to create DUR1,2. In the process, the allophanate hydrolyase domain (Dur 2) was inserted in the biotin-containing MccA. The Zygomycetes and Basidomycetes have only MccA (Fig. 1B in green), while the Hemiascomycetes are missing MccA (Fig. 1B). Split red-green and orange-green labels (Fig. 1B) indicate euascomycete species that contain both DUR1,2 and MccA. Schizosaccharomyces pombe does not contain either DUR1,2 or MccA. DUR1,2 likely originated prior to the split between the euascomycetes and hemiascomycetes and was subsequently lost from several euascomycete lineages, although the possibility for multiple independent origins cannot be eliminated.
There is ample precedent for metabolic dichotomies in the fungi. For instance, in lysine biosynthesis (Vogel, 1965) the two pathways for lysine biosynthesis are named after the intermediates that are characteristic of the paths, α-aminoadipic acid (AAA) and diaminopimelic acid (DAP). Euglenaceae and all of the fungi use the AAA pathway, whereas all other lysine prototrophs use the DAP pathway. No intermediates or enzymes are common to the two pathways (Vogel, 1965).
The distinct phylogenetic trees for urease (Fig. 1A) and urea amidolyase (Fig. 1B) raise three further questions regarding how and why those changes occurred. The first concerns why two plant pathogens, Gibberella zeae and Magnaportha grisea, retain both urease and urea amidolyase when plants recycle virtually all of their amino groups and thus do not excrete urea. The second concerns the energetics of biotinylated enzymes. Most eukaryotes have only four biotin-containing enzymes: pyruvate carboxylase, propionyl-CoA carboxylase, acetyl-CoA carboxylase, and methylcrotonyl-CoA carboxylase (Samols et al, 1988). Why do the hemiascomycetes use an energy-dependent, biotin-containing urea amidolyase system when the same overall reaction could be accomplished by the simpler urease? This question becomes even more germane when we consider that all strains of C. albicans are biotin auxotrophs (Odds, 1988), and it has long been known that 2 to 4 times as much biotin is required for maximum growth of S. cerevisiae on urea, allantoic acid, or allantoin as sole nitrogen sources (DiCarlo et al, 1953).
Loss of Ni2+/Co2+ enzymes
Our current thinking is that DUR1,2 allows the hemiascomycetes to retain urea as a nitrogen source while jettisoning their last Ni2+ -containing enzyme. As part of a comparative genomic analysis, Zhang et al (2009) examined sixty three fungal genomes for consensus/predictive sequences associated with Ni2+/Co2+ transporters and the use of nickel and cobalt by fungi. Among biometals, nickel and cobalt are considered together because they are used at particularly low levels and often share a transport system. For the cobalt-containing vitamin B12, three coenzyme B12-dependent enzymes: methionine synthase, methylmalonyl-CoA mutase, and the B12-dependent ribonucleotide reductase, were not found in any of the 63 fungal genomes (Zhang et al, 2009). Thus, in silico analysis concluded that the higher fungi as a group do not use Co2+ or any coenzyme B12-dependent enzymes. However, marine lower fungi belonging to the Phycomycetes may still have a functional requirement for cyanocobalamin and exhibit B12 deficiencies (Goldstein and Belsky, 1963).
The situation for nickel was more intriguing in that there was a precise dichotomy between the hemiascomycetes and the rest of the higher fungi. Zhang et al, (2009) screened for the nickel/cobalt transporter (Nic1p) and the Ni-dependent enzyme urease. Neither was present in any of the 24 hemiascomycete genomes examined, but both genes were present in all eight of the Basidiomycetes, both of the Schizosaccharomycetes, and 28 of the 29 Euascomycetes (Zhang et al, 2009). The exception was Aspergillus terreus ATTC 20542. Urease was the only Ni-dependent protein identified, and the taxonomic distribution of the nickel transporter and urease coincided exactly with that shown in Fig. 1A.
Thus, the hemiascomycetes do not have any Ni or Co dependent enzymes, thus avoiding the delicate balance of acquiring the necessary trace levels of Ni2+ and Co2+ without exceeding the threshold levels at which those transition metals become toxic. For instance, Mackay and Pateman (1980) described a mutant of A. nidulans for which, with urea as the sole nitrogen source, 0.1 mM Ni2+ was required but 1mM Ni2+ was toxic. By using DUR1,2 instead of urease, the Hemiascomycetes can eliminate Ni2+/Co2+ transporters and have two less essential transition metals. Also, humans do not utilize nickel for major metabolic processes, and nickel is generally viewed as a toxic or carcinogenic metal (Dosanjh and Michel, 2006). Switching to urea amidolyase would allow hemiascomycetes such as C. albicans to achieve urea degradation and kidney colonization in a nickel deficient human host.
Fusel Alcohols
The hemiascomycetes as a group have replaced methylcrotonyl-CoA carboxylase with urea amidolyase. Are there any phenotypes or negative consequences associated with this swap? MccA (EC 6.4.1.4) catalyzes the ATP-dependent carboxylation of 3-methylcrotonyl CoA from 3-methylglutaconyl CoA. It is certainly involved in leucine catabolism (Rodriguez et al (2004) and the production of fusel alcohols by yeasts may be a consequence of the loss of MccA. Fusel alcohols are derived from amino acid catabolism via a pathway proposed by Ehrlich (1907). For amino acids assimilated by the Ehrlich pathway (valine, leucine, isoleucine, methionine, and phenylalanine), after the initial transamination reaction the resulting α – κeto acids cannot be redirected into central metabolism and are instead decarboxylated, reduced, and excreted (Hazlewood et al, 2008). The suggestion that fusel alcohol production derives from the loss of MccA predicts that only the hemiascomycetes should be capable of fusel alcohol production. This prediction has been partially confirmed by Penalva and associates. They found that Δmcc strains of Aspergillius nidulans could not grow on leucine as the sole carbon source and accumulated 3-hydroxyisovaleric acid in the culture supernants. Although MccA is associated primarily with leucine catabolism (Rodriguez et al 2004), we note that Northern analysis of A. nidulans mycelia revealed that MccA and MccB transcription was elevated in media containing 30 mM of leucine, valine, isoleucine, methionine, or phenylalanine but not in any of the other four amino acids tested (Rodriquez et al, 2004).
Urea and Pathogenicity
Urea catabolism has relevance because urease is a virulence factor in at least two pathogenic fungi Cryptococcus neoformans (Cox et al, 2000) and Coccidioides immitis (Cole, 1997) and two bacteria Helicobacter pylori (Eaton et al, 1991) and Proteus mirabilis (Jones et al, 1990). Having cytoplasmic urea catabolism (urea amidolyase) permits urea-dependent signalling pathways related to fungal pathogenicity (Ghosh et al, 2009). We examined the role of arginine-induced germ tube formation in the escape of C. albicans from murine macrophages (Ghosh et al, 2009). Our studies link the work of Lorenz et al (2004), who showed that the genes for L-arginine biosynthesis were induced following internalization by macrophages, with that of Sims (1986) and Muhlschlegel’s group (Bahn and Mühlschlegel, 2006; Klengel et al, 2005) who showed that elevated CO2 triggers hyphal growth. We connected these two observations via the enzymes L-arginase (Car1p), which converts arginine to urea, and urea amidolyase (Dur1,2p) which produces CO2. At that time we created a dur1,2/dur1,2 mutant from a wild type parent (A72) and then reconstituted it. The dur1,2/dur1,2 mutant was unable to: grow on urea as the sole nitrogen source, stimulate germ tube formation in response to L-arginine or urea, or escape from the murine macrophage cell line RAW 264.7. These abilities were restored in the reconstituted strains (Ghosh et al, 2009). Finally, ongoing studies also show that DUR1,2 is a virulence factor for C. albicans (Navarathna and Roberts, unpublished).
Acknowledgments
We thank Donald G. Ahearn, Vadim Gladyshev, and anonymous reviewer #2 for helpful suggestions and discussions. This work was supported by the University of Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, the John C. and Nettie V. David Memorial Trust Fund, Ann L. Kelsall and the Farnesol and Candida albicans Research Fund, University of Nebraska Foundation, and the Intramural Research Program of the NIH, NCI, Center for Cancer Research (DDR).
References
- Bahn YS, Muhlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Cole GT. Ammonia production by Coccidiodes immitis and its posible significance to the host fungus interplay. In: Stevens D, Odds F, editors. Host-Fungus interplay. Bethesda: National foundation for infectious diseases; 1997. pp. 247–263. [Google Scholar]
- Cooper TG. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- Cooper TG, Lam C, Turoscy V. Structural analysis of the dur loci in S. cerevisiae: two domains of a single multifunctional gene. Genetics. 1980;94:555–580. doi: 10.1093/genetics/94.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastidar SG, Purandare NM, Desai SC. Growth requriements of Candida species. Indian J Exp Biol. 1967;5:228–232. [PubMed] [Google Scholar]
- DiCarlo FJ, Schultz AS, Kent AM. The mechanism of allantoin catabolism by yeast. Arch Biochem Biophys. 1953;44:468–474. doi: 10.1016/0003-9861(53)90064-2. [DOI] [PubMed] [Google Scholar]
- Dosanjh NS, Michel SLJ. Microbial nickel metalloregulation: NikRs gor nickel ions. Curr Opin Chem Biol. 2006;10:123–130. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Ber Dtsch Chem Ges. 1907;40:1027–1047. [Google Scholar]
- Galagan JE, Henn MR, Ma L-J, Cuomo CA, Birren B. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Research. 2005;15:1620–1631. doi: 10.1101/gr.3767105. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Inf Immun. 2009;77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Belsky M. B12 and B1 auxotrophy of lower marine phycomycetes. Arch Mikrobiol. 1963;47:161–163. [Google Scholar]
- Hazelwood LA, Daran JM, van Maris AJA, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Metab. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jones BD, Lockatell CV, Johnson DE, Warren JW, Mobley HL. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990;58:1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, Ekert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Mühlschlegel FA. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RK, Nickerson KW. Nutritional control of dimorphism in Ceratocystis ulmi. Exp Mycol. 1981;5:148–154. [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay EM, Pateman JA. Nickel requirement of a urease-deficient mutant in Aspergillus nidulans. J Gen Microbiol. 1980;116:249–251. doi: 10.1099/00221287-116-1-249. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and candidiasis. Tindall; Balliere: 1988. [Google Scholar]
- Rodriguez JM, Ruiz-Sala P, Ugarte M, Penalva MA. Fungal metabolic model for 3-methylcrotonyl-CoA carboxylase deficiency. J Biol Chem. 2004;279:4578–4587. doi: 10.1074/jbc.M310055200. [DOI] [PubMed] [Google Scholar]
- Roon RJ, Hampshire J, Levenberg B. Urea amidolyase. The involvement of biotin in urea cleavage. J Biol Chem. 1972;247:7539–7545. [PubMed] [Google Scholar]
- Roon RJ, Levenberg B. Urea amidolyase. I. Properties of the enzyme from Candida utilis. J Biol Chem. 1972;247:4107–4113. [PubMed] [Google Scholar]
- Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- Sentheshanmuganathan S, Nickerson WJ. Nutritional control of cellular form in Trigonopsis variabilis. J Gen Microbiol. 1962;27:437–449. doi: 10.1099/00221287-27-3-437. [DOI] [PubMed] [Google Scholar]
- Sims W. Effect of carbon dioxide on the growth and form of Candida albicans. J Med Microbiol. 1986;22:203–208. doi: 10.1099/00222615-22-3-203. [DOI] [PubMed] [Google Scholar]
- Vogel HJ. Lysine biosynthesis and evolution. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 25–42. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, colbalt, and vitamin B12 utilization. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]