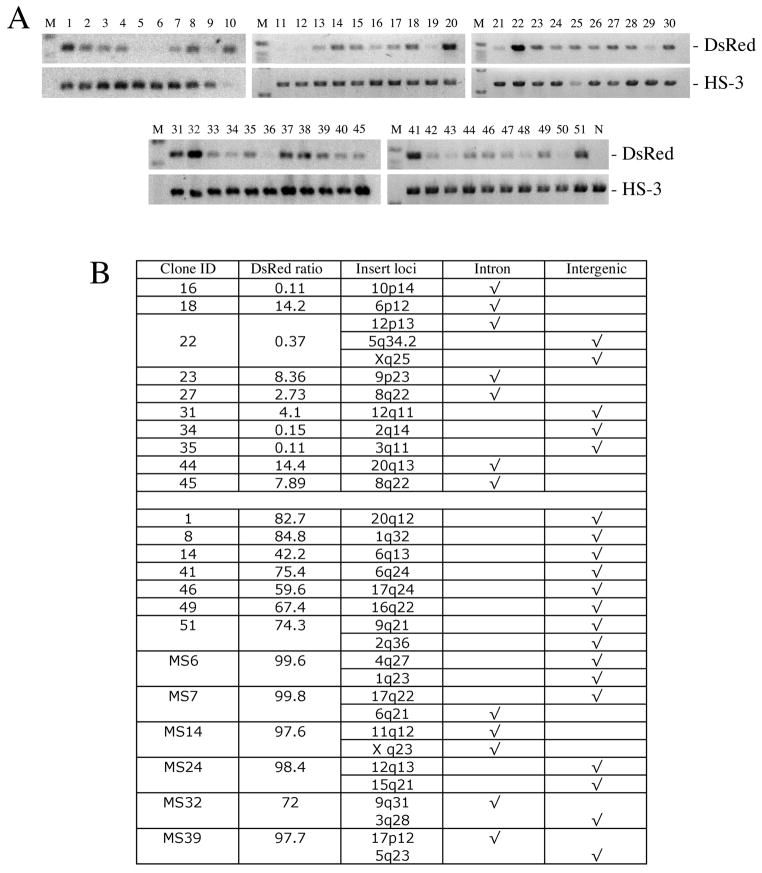
FIGURE 2.
Genomic copy number and chromosomal location of SB-IHK-DsRed insertions in individual K562 clones. (A) Genomic DNA isolated from single-cell clones was used as template for PCR using primers specific for DsRed and for the endogenous genomic HS3. (B) Determination of the SB-Tn insertion sites in cell clones. Using ligation-mediated nested PCR of the isolated genomic DNA, the insertion sites of the SB-IHK-DsRed were amplified and sequenced. NCBI BLAST analysis of the recovered flanking sequences was used to determine the genomic insertion sites in the K562 clones. The proportion of DsRed+ cells in each individual clones was determined by FACS analysis. Number of insertion loci identified by ligation-mediated PCR corresponded with the number estimated by semi-quantitative PCR in panel (A). MS prefix represents cell clones isolated by FACS machine whereas other cell clones were isolated manually.