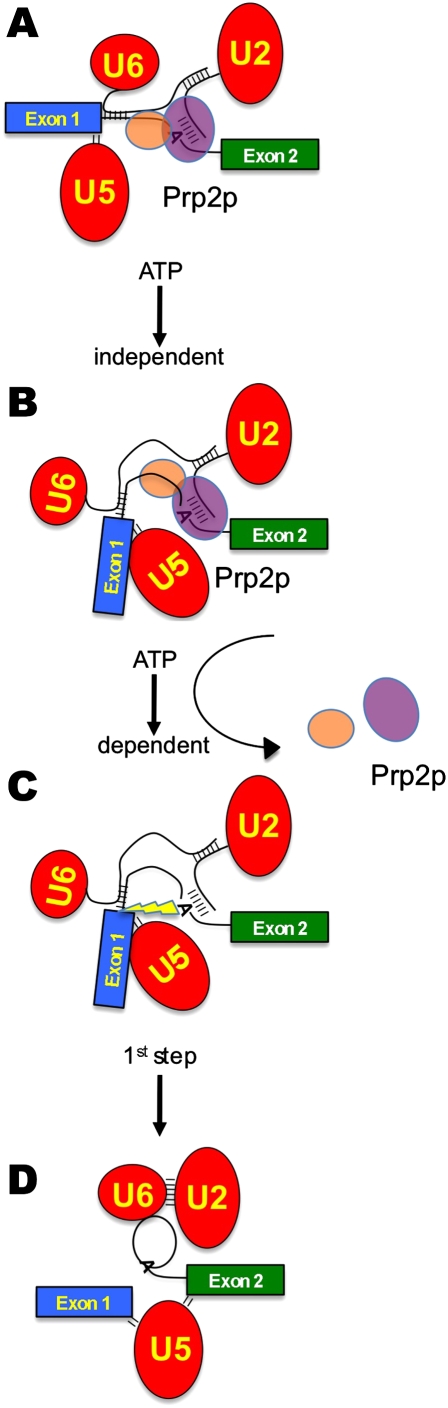
FIGURE 6.
Model for the activation of the first chemical step of pre-mRNA splicing. (A) The SF3b complex (purple oval) resides on the branchpoint/U2 RNA duplex, while SF3a (orange oval) resides just upstream of SF3b during spliceosome assembly and until the ATP-dependent action of Prp2p. The pre-mRNA is denoted by the green and blue exon boxes and the bottom RNA strand. snRNPs are shown as the red ovals with U2 and U6 snRNAs forming duplexes with each other and the pre-mRNA. U5 snRNP contacts the 5′ splice site. (B) After addition of Prp2p, an ATP-independent rearrangement concomitant with a structural rearrangement occurs. (C) Upon ATP hydrolysis by Prp2p, the SF3a and SF3b components are displaced from the pre-mRNA, uncovering the reactive branchpoint adenosine hydroxyl, which is now properly positioned for in-line attack of the 5′ splice site. (D) Transesterification releases exon1 and the lariat intermediate, formed by the 2′–5′ linkage between the branchpoint and the 5′ splice site.