Abstract
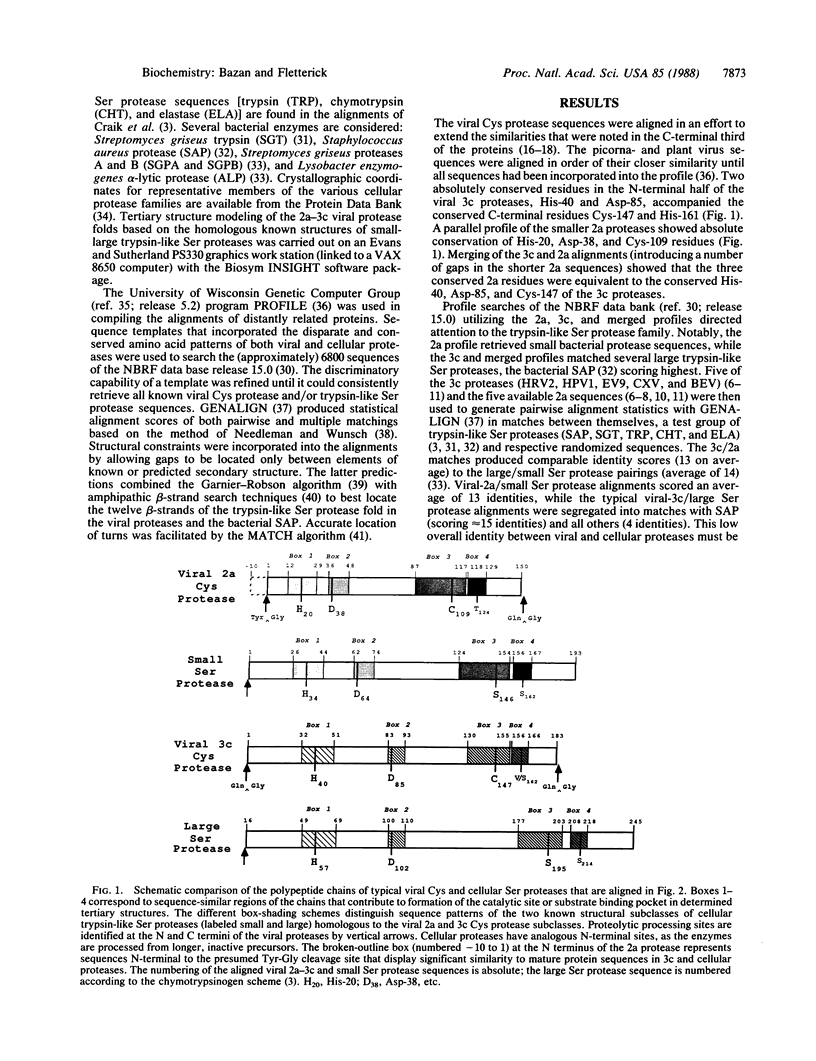
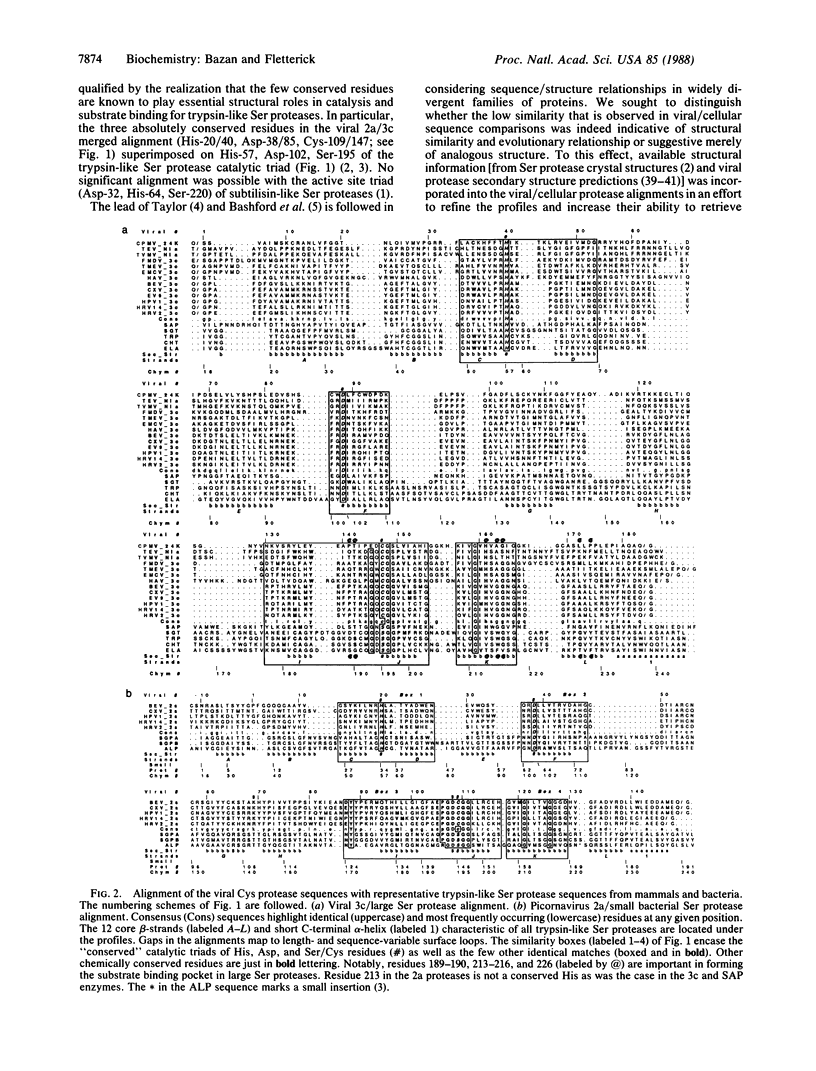
Proteases that are encoded by animal picornaviruses and plant como- and potyviruses form a related group of cysteine-active-center enzymes that are essential for virus maturation. We show that these proteins are homologous to the family of trypsin-like serine proteases. In our model, the active-site nucleophile of the trypsin catalytic triad, Ser-195, is changed to a Cys residue in these viral proteases. The other two residues of the triad, His-57 and Asp-102, are otherwise absolutely conserved in all the viral protease sequences. Secondary structure analysis of aligned sequences suggests the location of the component strands of the twin beta-barrel trypsin fold in the viral proteases. Unexpectedly, the 2a and 3c subclasses of viral cysteine proteases are, respectively, homologous to the small and large structural subclasses of trypsin-like serine proteases. This classification allows the molecular mapping of residues from viral sequences onto related tertiary structures; we precisely identify amino acids that are strong determinants of specificity for both small and large viral cysteine proteases.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Garavito R. M., Eventoff W., Rossmann M. G., Brändén C. I. Similarities in active center geometries of zinc-containing enzymes, proteases and dehydrogenases. J Mol Biol. 1978 Dec 5;126(2):141–158. doi: 10.1016/0022-2836(78)90356-x. [DOI] [PubMed] [Google Scholar]
- Argos P., Kamer G., Nicklin M. J., Wimmer E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 1984 Sep 25;12(18):7251–7267. doi: 10.1093/nar/12.18.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D., Chothia C., Lesk A. M. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987 Jul 5;196(1):199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Carrington J. C., Dougherty W. G. Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J Virol. 1987 Aug;61(8):2540–2548. doi: 10.1128/jvi.61.8.2540-2548.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen F. E., Abarbanel R. M., Kuntz I. D., Fletterick R. J. Turn prediction in proteins using a pattern-matching approach. Biochemistry. 1986 Jan 14;25(1):266–275. doi: 10.1021/bi00349a037. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Rutter W. J., Fletterick R. Splice junctions: association with variation in protein structure. Science. 1983 Jun 10;220(4602):1125–1129. doi: 10.1126/science.6344214. [DOI] [PubMed] [Google Scholar]
- Delbaere L. T., Brayer G. D., James M. N. The 2.8 A resolution structure of Streptomyces griseus protease B and its homology with alpha-chymotrypsin and Streptomyces griseus protease A. Can J Biochem. 1979 Feb;57(2):135–144. doi: 10.1139/o79-017. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier L. L., Franklin K. M., Shahabuddin M., Hellmann G. M., Overmeyer J. H., Hiremath S. T., Siaw M. F., Lomonossoff G. P., Shaw J. G., Rhoads R. E. The nucleotide sequence of tobacco vein mottling virus RNA. Nucleic Acids Res. 1986 Jul 11;14(13):5417–5430. doi: 10.1093/nar/14.13.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau G. R. The primary structure of staphylococcal protease. Can J Biochem. 1978 Jun;56(6):534–544. doi: 10.1139/o78-082. [DOI] [PubMed] [Google Scholar]
- Earle J. A., Skuce R. A., Fleming C. S., Hoey E. M., Martin S. J. The complete nucleotide sequence of a bovine enterovirus. J Gen Virol. 1988 Feb;69(Pt 2):253–263. doi: 10.1099/0022-1317-69-2-253. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J., Stroud R. M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Jan;81(1):155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forss S., Strebel K., Beck E., Schaller H. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 1984 Aug 24;12(16):6587–6601. doi: 10.1093/nar/12.16.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H., Leunissen J., Goldbach R., Lomonossoff G., Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984 Apr;3(4):855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. A., Schrijvers L., Tan A., Vos P., Wellink J., Goldbach R. Proteolytic activity of the cowpea mosaic virus encoded 24K protein synthesized in Escherichia coli. Virology. 1987 Jul;159(1):67–75. doi: 10.1016/0042-6822(87)90348-5. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P. Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families. FEBS Lett. 1986 Jan 6;194(2):253–257. doi: 10.1016/0014-5793(86)80095-3. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Homyak M., Edenfield J., Eisenberg D. Profile scanning for three-dimensional structural patterns in protein sequences. Comput Appl Biosci. 1988 Mar;4(1):61–66. doi: 10.1093/bioinformatics/4.1.61. [DOI] [PubMed] [Google Scholar]
- Hanecak R., Semler B. L., Ariga H., Anderson C. W., Wimmer E. Expression of a cloned gene segment of poliovirus in E. coli: evidence for autocatalytic production of the viral proteinase. Cell. 1984 Jul;37(3):1063–1073. doi: 10.1016/0092-8674(84)90441-0. [DOI] [PubMed] [Google Scholar]
- Higaki J. N., Gibson B. W., Craik C. S. Evolution of catalysis in the serine proteases. Cold Spring Harb Symp Quant Biol. 1987;52:615–621. doi: 10.1101/sqb.1987.052.01.070. [DOI] [PubMed] [Google Scholar]
- Ivanoff L. A., Towatari T., Ray J., Korant B. D., Petteway S. R., Jr Expression and site-specific mutagenesis of the poliovirus 3C protease in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5392–5396. doi: 10.1073/pnas.83.15.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant B. D., Brzin J., Turk V. Cystatin, a protein inhibitor of cysteine proteases alters viral protein cleavages in infected human cells. Biochem Biophys Res Commun. 1985 Mar 29;127(3):1072–1076. doi: 10.1016/s0006-291x(85)80054-1. [DOI] [PubMed] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- König H., Rosenwirth B. Purification and partial characterization of poliovirus protease 2A by means of a functional assay. J Virol. 1988 Apr;62(4):1243–1250. doi: 10.1128/jvi.62.4.1243-1250.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. M., Stålhandske P. O., Pettersson U. Genome of coxsackievirus B3. Virology. 1987 Jan;156(1):50–63. doi: 10.1016/0042-6822(87)90435-1. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Shanks M. The nucleotide sequence of cowpea mosaic virus B RNA. EMBO J. 1983;2(12):2253–2258. doi: 10.1002/j.1460-2075.1983.tb01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M. A flexible multiple sequence alignment program. Nucleic Acids Res. 1988 Mar 11;16(5):1683–1691. doi: 10.1093/nar/16.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarian R., Caput D., Gee W., Potter S. J., Renard A., Merryweather J., Van Nest G., Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985 May;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neurath H. Evolution of proteolytic enzymes. Science. 1984 Apr 27;224(4647):350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- Palmenberg A. C., Kirby E. M., Janda M. R., Drake N. L., Duke G. M., Potratz K. F., Collett M. S. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 1984 Mar 26;12(6):2969–2985. doi: 10.1093/nar/12.6.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. A structural model for the retroviral proteases. Nature. 1987 Sep 24;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Pevear D. C., Calenoff M., Rozhon E., Lipton H. L. Analysis of the complete nucleotide sequence of the picornavirus Theiler's murine encephalomyelitis virus indicates that it is closely related to cardioviruses. J Virol. 1987 May;61(5):1507–1516. doi: 10.1128/jvi.61.5.1507-1516.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. J., Brayer G. D., Jurásek L., James M. N. Critical evaluation of comparative model building of Streptomyces griseus trypsin. Biochemistry. 1984 Dec 18;23(26):6570–6575. doi: 10.1021/bi00321a045. [DOI] [PubMed] [Google Scholar]
- Sidman K. E., George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1988 Mar 11;16(5):1869–1871. doi: 10.1093/nar/16.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielecki A. R., Hendrickson W. A., Broughton C. G., Delbaere L. T., Brayer G. D., James M. N. Protein structure refinement: Streptomyces griseus serine protease A at 1.8 A resolution. J Mol Biol. 1979 Nov 15;134(4):781–804. doi: 10.1016/0022-2836(79)90486-8. [DOI] [PubMed] [Google Scholar]
- Skern T., Sommergruber W., Blaas D., Gruendler P., Fraundorfer F., Pieler C., Fogy I., Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985 Mar 25;13(6):2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Minor P. D., Almond J. W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984 Oct 25;12(20):7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Reeve P., Minor P. D., Schild G. C., Almond J. W. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1539–1543. doi: 10.1073/pnas.81.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R. Identification of protein sequence homology by consensus template alignment. J Mol Biol. 1986 Mar 20;188(2):233–258. doi: 10.1016/0022-2836(86)90308-6. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Wellink J., van Kammen A. Proteases involved in the processing of viral polyproteins. Brief review. Arch Virol. 1988;98(1-2):1–26. doi: 10.1007/BF01321002. [DOI] [PubMed] [Google Scholar]
- Werner G., Rosenwirth B., Bauer E., Seifert J. M., Werner F. J., Besemer J. Molecular cloning and sequence determination of the genomic regions encoding protease and genome-linked protein of three picornaviruses. J Virol. 1986 Mar;57(3):1084–1093. doi: 10.1128/jvi.57.3.1084-1093.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosawa H., Ojima S., Ishii S. Thioltrypsin. Chemical transformation of the active-site serine residue of Streptomyces griseus trypsin to a cysteine residue. J Biochem. 1977 Sep;82(3):869–876. doi: 10.1093/oxfordjournals.jbchem.a131763. [DOI] [PubMed] [Google Scholar]



