Abstract
Cytoplasmic dynein is an important motor that drives all minus-end directed movement along microtubules. Dynein is a complex motor whose processive motion is driven by ATP-hydrolysis. Dynein's run length has been measured to be several millimetres with typical velocities in the order of a few nanometres per second. Therefore, the average time between steps is a fraction of a second. When this time scale is compared with typical time scales for protein side chain and backbone movements (~10−9 s and ~10−5 s, respectively), it becomes clear that a multi-timescale modelling approach is required to understand energy transduction in this protein. Here, we review recent efforts to use computational and mathematical modelling to understand various aspects of dynein's chemomechanical cycle. First, we describe a structural model of dynein's motor unit showing a heptameric organization of the motor subunits. Second, we describe our molecular dynamics simulations of the motor unit that are used to investigate the dynamics of the various motor domains. Third, we present a kinetic model of the coordination between the two dynein heads. Lastly, we investigate the various potential geometries of the dimer during its hydrolytic and stepping cycle.
1. Introduction
Cytoplasmic dynein drives nearly all minus-end directed microtubule-based movement in eukaryotic cells. Dynein's function specifically includes spindle formation and chromosome segregation and the transport of numerous cargoes including viruses, RNA, signalling molecules, and organelles.3 The loss of dynein function has been associated with major human diseases such as schizophrenia, lissencephaly, and motor neuron degeneration.4 Thus, understanding the molecular basis of dynein function is essential to our understanding of fundamental biological processes and many human diseases.
Dyneins are multi-component complexes that are constructed around one to three heavy chains that contain the ATPase and motor activities. Dynein are members of the ancient AAA+ (ATPases associated with diverse cellular activities) family of ATPases that includes a wide variety of proteins.2,5,6 The heavy chain is composed of a tail that binds to various cargos and other intermediate proteins, a microtubule binding stalk, and a motor unit that binds and hydrolyzes ATP and putatively is the site for force production (Fig. 1A).
Fig. 1.
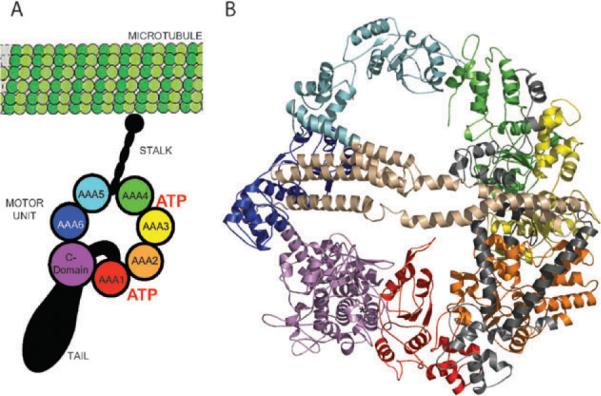
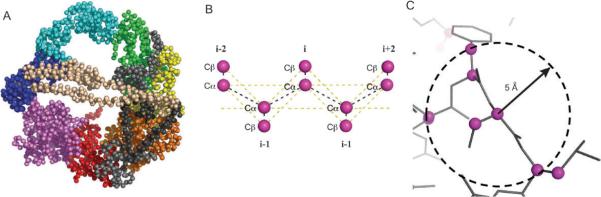
Model of the cytolasmic dynein motor head. (A) Schematic of the dynein showing its three major structural components: a tail involved in carrying cargoes, binding to intermediate and light chains, and oligomerization with other dynein heads; an ATP-hydrolysing motor domain; and a microtubule binding stalk. (B) Theoretical model of the motor unit constructed from homology models of the six AAA+ units, C-domain, and the interdomain regions (IDRs). The model is coloured according to the schematic. (Panel B is adapted from ref. 2.)
The stepping mechanism of single dynein has been explored by many groups using single molecule assays. Studies of bead movement driven by cytoplasmic dynein in vitro suggest that single molecule dynein molecules are processive. That is, single molecules of dynein are capable of taking multiple steps along the microtubule track without detaching.7–11 The mechanisms of processivity of Kinesin 1 and Myosin V have been well studied, and it has been shown that nucleotide-driven conformational changes of their mechanical elements power the hand-over-hand stepping of their two identical motor domains.11,12
In contrast, the mechanism of processivity in dynein is much less well understood, and dynein's distinct evolutionary origin and structural features suggest that its mechanism differs considerably from other molecular motors. In a recent single-molecule analysis, Vale et al. showed that the processivity of cytoplasmic dynein require the coordination of its two heads.13 They also found the minimal molecular components that displayed processive motion. This minimal unit consisted of the two motor domains and the immediately adjacent linker, thus none of the known dynein-associated subunits in the yeast genome are required for processivity.13
Using dynein molecules labelled with quantum-dots, it was shown that dynein takes both small (~8 nm) and large (12–24 nm) step sizes with occasional backward stepping, which is rarely observed in Kinesin 1 or Myosin V. Moreover, analysis of the dwell times between steps suggests that the dynein dimer consumes one ATP per step (kcat ~ 16 Pi s−1).13 However, despite these recent advances in single-molecule measurements, a detailed structural and mechanistic understanding of force generation by dynein remains elusive.
Here, we review various computational approaches for understanding the chemomechanical mechanism of cytoplasmic dynein's processive motion. First, we describe our effort in modelling the structure of the dynein motor unit, which is the force-producing domain of the molecule. Second, we describe our molecular dynamics simulations of the motor unit to elucidate the domain movements within the motor unit that may be associated with its force production. Third, we develop a kinetic model for the coordinated stepping of the motor. Lastly, we explore the various dimerization geometries of the motor unit. By using this multiscale approach, our goal is to gain a comprehensive understanding of dynein function.
2. Modelling the structure of the cytoplasmic dynein motor unit
To gain insight into the mechanism of force production, we constructed a structural model of the cytoplasmic dynein motor unit using homology to other AAA units.2 Sequence analysis of the dynein's motor unit indicates that it consists of six concatenated AAA subunits, an extended stalk that contains a microtubule binding domain, and a C-terminal domain that is twice the size of an AAA subunit (Fig. 1A).2,5,6
Electron microscopy (EM) reconstruction of the dynein motor unit at ~25 Å resolution form negatively stained Dictyostelium discoideum shows a complex of seven densities arranged in an asymmetric ring with “smooth” and “rough” edges.14 To determine the tertiary organization of the various domains, we fit the model structures into the EM density.2 From the results of Koonce et al., we assume that: (1) the seven lobe densities of the motor correspond to the six AAA+ units and the C-domain. More specifically, we assumed that the smooth side consists of the more conserved AAA1-AAA4 domains while the rough side of the motor consists of AAA5, AAA6, and C-domain.14 Because the sequences of these domains are less conserved, they are not expected to follow the configuration of homomeric AAA complexes, and therefore break the symmetry of the motor ring.2,14 To preserve the functionally relevant interactions between the domains AAA1-AAA4 and to construct a regular tetramer for this portion of the motor, we further assume that: (2) the orientation of AAA1-AAA4 units follows the orientation of the σ54 RNA polymerase activator NtrCl (PDB ID: 1NY6).15 This polymerase is a AAA complex that forms a homo-heptameric unit, thus preserving the correct interdomain interaction between units of AAA that form into a heptameric unit. This assumption satisfies the observation that the active sites of the first four AAA units are conserved and are still able to bind ATP although at different rates, which suggests that they should still preserve the functional interfaces between the AAA units.
Dynein, despite consisting of multiple domains, comes from a single polypeptide chain. In terms of primary structure, there exists linker regions between the AAA and C-domains. These regions range in length from 79 to 231 residues long, and consists primarily of helices. The most interesting of these linkers is interdomain 4 (IDR4) found between AAA5 and AAA6. This is the largest interdomain linker, which has a size (~200 residues) almost about the size of an AAA unit.2 Coiled-coil prediction algorithms assign a coiled-coil structure in the AAA5-ID4 sequence, although the length of this predicted coiled-coil region varies from one specie to another.2 Interestingly, in one of the EM maps there are densities on the faces of the ring. One of these densities forms a long arc across the diameter of the motor unit and is suggestive of a coiled-coil structure. Based on these results and observations, we postulate that the interdomain linker IDR4 is a coiled-coil structure that spans the diameter of the motor unit on one side, and that the tail of the motor unit attaches to the motor domain through the opposite side.
We constrcted models of the AAA+ domains from homology to known AAA+ domains. These AAA domains consist of the conserved α/β Rossman fold subdomain and an α-helical globular subunit.2 Despite a low (<20%) sequence identity, the model domains fold to within 2 Å root mean square deviation (RMSD). We also constructed a putative model of the C-terminal domain that consists of the first 290 residues folding into an all-α domain, while the remaining residues consists of five β-strands terminating in a helix.2 To orient AAA5, AAA6, and the C-domain, we fit them separately into their individual corresponding densities (Fig. 1B).
We also constructed models for the linker regions between the AAA units. Searching for homologues to this region likewise returned structures that were coiled-coil. Thus, we modelled ID4 region as a coiled-coil that fits into the over arching density on the face of the ring.2
The predicted coiled-coil region that spans across the motor unit is no inconsistent with the suggestion that the tail connects to the motor by coming through the face of the motor. It should be noted that in the EM reconstruction studies that both faces of the motor unit exhibit residual densities.14 While one of the density spans the motor, there is also a density that runs across half of the motor, which may be the remnant of the tail.
3. Investigation of motor unit dynamics using molecular dynamics simulation
To investigate the dynamics of the dynein motor unit, we performed molecular dynamics simulation using a simplified structural model of the protein (Fig. 2).16 The simplified protein model uses two beads per residue that are derived from the Cα and Cβ carbon atoms of each residue (see Appendix A). Another level of simplification is applied to approximate the interaction between particles. The model used Go-type interactions whereby particles that are within a cutoff distance (~5 Å) are assigned an attractive interaction while those beyond the cutoff are assigned a repulsive interaction.17–23 To calculate the evolution of the system, we use discrete molecular dynamics whereby the interaction potentials are approximated using square wells. By discretizing the potentials, the evolution of the system is driven by the ballistic collision between particles as opposed to time integration of Newtonian equations of motion as performed in traditional molecular dynamics (see Appendix B).18 This type of a simplified protein model and interactions between the protein particles has been used to elucidate the dynamics of another large protein, vinculin,24 and in reproducing the experimentally observable two-state folding of the src-SH3 protein.25,26
Fig. 2.
Coarse-grained model of the dynein motor head. (A) A two-bead-per-residue model of the dynein head. Each residue is represented by their Cα and Cβ atoms. (B) Effective interactions defined in the two-bead model. Yellow lines show non-covalent interactions while the black lines show covalent interactions. (C) The interaction within each head is determined by the Go-model whereby neighbouring atoms are assigned an attractive interaction while those that are far from each other are assigned a repulsive interaction.
Using the simplified model described above, we extensively characterized the dynamics of dynein. First, we performed discrete molecular dynamics (DMD) simulations for 106 time units (approximately a few milliseconds) with initial temperatures from T = 0.1 ε/kBT to T = 2.0 ε/kBT (see Appendix A for units).18,27 These equilibrium simulations allowed us to determine the thermal denaturation curve of the dynein head and the melting temperature. Using the observation that the native state of the protein is slightly below the melting temperature, we then performed molecular dynamics simulations near the identified melting point.
To quantify the fluctuations of all the domains, we calculated the per residue root mean square deviation (RMSD) with respect to the initial structure. The average fluctuations of residues within a particular domain are shown in Fig. 2. We found the “rough” side of the motor (Fig. 3) composed of AAA5, IDR4, AAA6, and C-domain exhibits the largest fluctuations, whereas the “smooth” side, which is composed of the AAA1 to AAA4 is a more ccompact structure. Interestingly, the ATP-binding domains are located on the smooth side, suggesting that only minor conformational changes in the catalytic binding pocket are induced upon hydrolysis or product release, however, these conformations are then propagated to and amplified by the rough side of the motor.
Fig. 3.
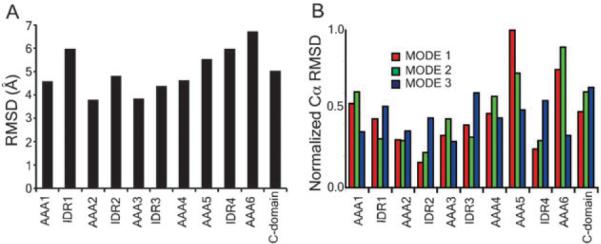
Backbone fluctuations of the motor unit. (A) The per residue root-mean-square deviation with respect to the starting conformation was calculated over the equilibrium simulation run. (B) Backbone fluctuations calculated using the normal mode analysis. Shown are the lowest three modes of the protein. The observation of greatest variability in domains AAA5, AAA6, and the C-domain is in agreement with reconstructed 3D EM structures of the motor unit in three distinct stalk conformations. In the three stalk positions, the side formed by AAA5, AAA6, and C domain exhibit the largest variation. (Panel B is adapted from ref. 2.)
These results from long DMD simulations are in agreement with our analysis of the lowest energy normal modes of dynein.2 Normal mode analysis has been shown to accurately identify the structural sites that function as pivots and, therefore, can be used to infer global motions of large molecular complexes.28,29 From Fig. 3B, it is evident that the most mobile domains are A5, A6, and the C-domain, whereas domains A1 to A4 form a more compact structure. These observations are quantified in Fig. 3B, which lists the RMSD of the Cα atoms of each subunit for the first three normal modes. In all the three lowest modes, the motion of the catalytic site is smaller compared with the dominant motion in that mode.2
These observations from molecular dynamics and normal mode analysis suggest that during the mechanochemical events leading to force production, only minor conformational changes occur in the catalytic site upon binding or release of ATP or ADP. However, this small conformational change is amplified in the rest of the motor domains. More importantly, in 3D EM reconstructions of the isolated dynein motor unit with stalk positioned at 0°, 25°, and 45° relative to vertical axis show greatest variation in the densities of AAA5, and AAA6 (Fig. 3C).14
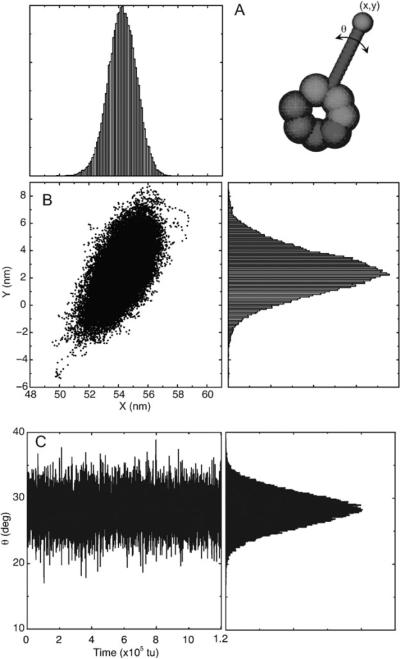
One major difference between dynein and the other cytoskeletal motors myosin and kinesin is that dynein's microtubule binding domain is located ~15 nm from the motor unit and ~30 nm from the primary catalytic site.30,31 Myosin and kinesin on the other hand have integrated their sites for catalysis and actin- or microtubule-binding.32,33 Thus, major structural difference suggests that dynein's mechanism of modulating its binding/unbinding to the MT is very different from that of either myosin or kinesin. To investigate how the dynamics within the motor unit are transmitted to the microtubule binding stalk, we constructed a geometric stalk as the perpendicular bisector between the centers of mass of the AAA4 and AAA5 unit, from which the microtubule binding stalk emanates. This geometric bisector is assumed to be ~15 A long which is the length of dynein's MT binding stalk as measured from reconstructed EM map images. Shown in Fig. 4 is the trace of the stalk tip as projected on the plane of the motor unit, which indicates that the potential fluctuations of the stalk tip is approximately ~5 nm. We likewise calculated the angular deviations of the projected stalk (Fig. 4B). The stalk equilibrium deviations is ~3°. This angular deviation of the theoretical stalk is enough to cause the sliding of between the strands of the coiled-coil stalk that connects the microtubule-binding domain to the motor unit. The sliding between the coiled-coiled strands have been shown to modulate the binding affinity of MT-binding domain to the microtubule.34,35 Thus, the intrinsic fluctuations of the motor unit is enough to modulate the dynein's affinity to the microtubule.
Fig. 4.
Motion of a putative stalk during equilibrium molecular dynamics simulations. (A) The stalk is constructed as the perpendicular bisector of the centers of mass of the AAA4 and AAA5 domains. (B) Trace of the stalk tip positions. (C) The angular displacement is measured relative to the initial position of the relative stalk.
4. Kinetic models and stochastic simulations of dynein function
A mechanistic understanding of the mechanochemical cycle that drives motor function is an important goal in the field of motor proteins. Because of dynein's large size, molecular dynamics simulations are unable to capture the time scales relevant to its processive motion. However, coarse-grained stochastic models have proven to be very useful tool for analyzing and interpreting experimental data, such as velocity, processivity, dwell-times and step size distributions, for other motor proteins. For example, in combination with MD simulations, structural analysis and experimental measurements, stochastic models have been essential for understanding the mechanochemical cycle of myosin V and more recently conventional kinesin.36–38
In their in vitro motility experiments Shima et al. showed that at least four single-headed dynein molecules are required to generate continuous sliding of microtubules comparable to that of a single two-headed full-length dynein.39,40 This result supports the conjecture that head-head coordination is required for the processive motion of cytoplasmic dynein. Although it is clear that for processive stepping the attachment-detachment cycles of the two heads need to be out of phase, coordination is also necessary to achieve fast head-over-head stepping for long run lengths. A trivial form of coordination is achieved if the mechanochemical cycle contains a single rate-limiting step, such as ADP release from a head strongly bound to the microtubule. In this case, the motor dwells most of the time in the state in which both heads contain ADP and are bound to the microtubule. Eventually one of the heads releases ADP, goes quickly through its full cycle, and returns back to the bound ADP state, before the other head has a chance to release ADP. Clearly such a motor would be very processive, but also very slow. Its stepping would contain both head-over-head and inch-worming motions. Much more effective stepping in terms of the velocity and the processivity is achieved by a non-trivial coordination that provides out-of-phase cycling of not only attachment and detachment from the microtubule but also other mechano-chemical transitions, such as ATP hydrolysis and the power stroke.
Because monomeric dynein sufficient to function as a motor, a single head has to synchronize two mechanical cycles (attachment/detachment and pre-stroke/post-stroke) with a biochemical cycle (ATP binding/hydrolysis/product release). Based on kinetic studies of single-headed dynein, we adopt the following model: (1) the cycle of a single-headed dynein consists of the following sequence of events: ATP binding, detachment, post-stroke to pre-stroke transition (recovery), hydrolysis, phosphate release, attachment, power-stroke, and ADP release. We also assume that: (2) for a single non-interacting head the rates of both the power-stroke and ADP release are slow with respect to the other rates in the cycle.
Although head-head coordination is achieved by physical interactions,41 these interactions manifest themselves by modulating kinetic characteristics of one head dependent upon the biochemomechanical state of the other. Given the kinetics of a single-headed dynein one can explore the full network of all mechanochemical states of the two-headed motor. In this case, the mechanochemical cycle of the motor is represented by a subset of states (pathway) isolated from the rest states by a unique set of kinetic rules. Then, plausible pathways and, hence, a plausible coordination scheme (s), can be chosen based on the restrictions imposed by the experimentally measured characteristics of the motor's performance, without an a priori knowledge of the physical nature of the heads interactions (for details see ref. 42).
Following this philosophy, we found that the minimal head-to-head coordination required for highly processive and sufficiently fast head-over-head stepping includes: (A) the acceleration of the ADP release rate by formation of a compact conformation in which both heads are bound to the microtubule 8.2 nm apart and (B) the acceleration of the pre-stroke to post-stroke transition of a bound head by the detachment of the other one.
Under these two kinetic rules the dominant pathway consists of the following steps. After detachment of the trailing head A (upon ATP binding) the bound head B in the ADP state rapidly performs the power-stroke; the free head A then goes relatively fast trough a series of transitions including recovery, ATP hydrolysis, phosphate release and attachment to the microtubule, which accelerates ADP release from the head B and completes the half cycle resulting in head A moving in front of head B. It is important to note that the asymmetric release of ADP results from the two heads being in distinct mechanical states. This effect is achieved automatically because the leading head with ADP bound is in the pre-stroke conformation. According to the single-headed model,42,43 in order to release ADP this head has to perform the power-stroke, which is a slow transition until the rear head detaches. In other words, it is essential that even if the heads turn out to be in the same chemical state when both are bound to the microtubule, they must be in different mechanical states to ensure that they stay out of phase.42
Since we did not assume any other interactions between the bound and free head, our network allows for two branching points in the pathway: (1) the recovery of the free head can occur before or after the power-stroke of the bound head, and (2) if recovery is first, hydrolysis by the free head can occur before or after the power-stroke of the bound head.
Having established the order of transitions in the mechano-chemical cycle of two-headed dynein, one can compute the behaviour of the motor in different parameter regimes. Specifically, we explored the sensitivity of the motors speed and run-length to the changes in the nucleotide concentrations and in the rates modulated by the head-head coordination. The additional ATP-binding sites in the AAA2-AAA4 domains are thought to play a regulatory role in motor function and, thus, potentially in the coordination of the two heads.44,45 Cho et al. used mutagenesis to disrupt ATP hydrolysis in these sites.46 Comparing their experimental results with the performance of our model, we found that the increase in the run length without a significant change in the velocity observed when the AAA4 site is disrupted is captured by the model when the rate of ADP release from the primary site of the attached head in the non-compact conformation is reduced. Similarly, the decrease of both the run length and the velocity, as well as force generation observed when the AAA3 site is disrupted can be attributed to either a reduction in the rate of the power-stroke of the attached head in the non-compact conformation or a reduction in the affinity of the heads for the microtubule.
Finally, our two-headed model can be used as a starting point for an extended model accounting for the rich stepping behaviour of dynein, including wide step-size distribution, frequent back steps, diffusive motion, etc. The transition to these types of stepping behaviour can be associated with an occasional disruption of the compact conformation, which is likely essential for the tight head-head coordination. We speculate that the ability of dynein to switch between stepping patterns is a mechanism which allows the motor to avoid obstacles and navigate in the crowded cellular environment.47 Within our model the probable switch point if the free head attaches in a position that does not produce a compact confirmation. For example, if this head attaches more than one microtubule period (8.2 nm) away from the bound head. Depending on the choice of kinetic parameters for the split at the switch point the compact conformation can be restored on the next half cycle or later. Trafficking characteristics of the motors generated by such extended models is an ongoing research project.48
5. Power stroke and stepping geometry
Linear molecular motors under fixed conditions transduce chemical energy into directed mechanical motion. Such directionality is determined by the intrinsic asymmetry of the polymer track. However, the fact that dynein's heads bind to the microtubule in a specific orientation is not enough to explain processive head-over-head stepping in one direction. The widely accepted view is that the mechanochemical cycle of the motor unit contains a conformational change that biases the diffusive motion of the detached head in the forward direction—the so called `power-stroke'. Dynein's pre-stroke and post-stroke conformations have been visualized by Burgess et al. using electron microscopy.1 However, these results and subsequent experimental studies35,43 have not produced a consistent picture of force generation by dynein.
Example 1. In their model, Burgess et al. considered a single-headed dynein with a fixed tail, so that during the power-stroke the AAA+ ring rotates around its axis and moves the tip of the stalk in the direction of the plus end of the microtubule. This model is indeed consistent with the plus end directed motion of the microtubule observed in sliding assays. However, if one adopts this model for cytoplasmic dynein and fixes the stalk instead of the tail, then the motion of the tail's tip (cargo-binding domain) will again be in the direction of the microtubule's plus end, which is not consistent with the minus-end directed motion of the motor. One way to reconcile the discrepancy is to assume that the angle between the stalk and the microtubule changes during the power-stroke.
Example 2. Mizuno et al. have interpreted the power-stroke in Burges model as motion of the AAA+ ring, in which the ring-stalk complex leans forward (toward minus end) from a perpendicular orientation in the pre-stroke state. Then based on their own cryo-EM visualization, they proposed an alternative scenario, in which the ring-stalk complex moves from a backward leaning position to a perpendicular orientation in the post-stroke state. In both cases the ring moves toward the minus end of the MT, but the orientation of AAA subunits of the ring is reversed.
Example 3. In another study, Carter et al. considered two alternative models of force generation.35 First, they interpreted Burgess's results by associating the power-stroke only with the motion of the linker rather than the tail (presumably taking into account the flexibility of the N terminus of the dynein heavy chain). From this view, the tail-linker junction moves around the ring as expected toward the minus end even with a fixed orientation of the ring-stalk complex during the power stroke. Then, the authors suggested that the force generated by the power-stroke is directed strictly toward the microtubule-binding domain along the stalk with a positive projection of the force toward the minus end of MT while the ring-stalk complex is leaning backward. This geometry was proposed by the authors to explain their observation that a dynein construct with a flipped orientation of the AAA+ ring with respect to the microtubule binding domain maintains minus end directed stepping.
It is important to note that all these seemingly different models only consider an isolated dynein head independent of its partner. Therefore, these models do not actually explain how the power-stroke fits into the coordinated head-over-head stepping of the motor and how this conformational change biases forward progression of the heads. To address this issue, we use simple geometrical considerations and our two-headed kinetic model to propose a detailed model for dynein stepping in agreement with the experimental observations mentioned above.
5.1 Motor geometry during the kinetic cycle
To begin with we ignore the flexibility of both the tail and the stalk, and arrange two heavy chains in the pre-stroke and post-stroke conformations (adopted from Fig. 4 in Burgess et al.) with linked tails and the stalks attached to the microtubule 8.2 nm apart. According to our two-headed model, when both heads are strongly bound to the microtubule, the trailing head should be in post-stroke conformation (Apo-state) while the leading head is in the pre-stroke state. In this geometry the AAA+ ring of the trailing head is positioned in front of the leading head's ring. Here we assume that the post-stroke head holds most of the external load, so that the force line goes through a point close to where the trailing head binds the microtubule and through the tails junction (Fig. 6).
Fig. 6.
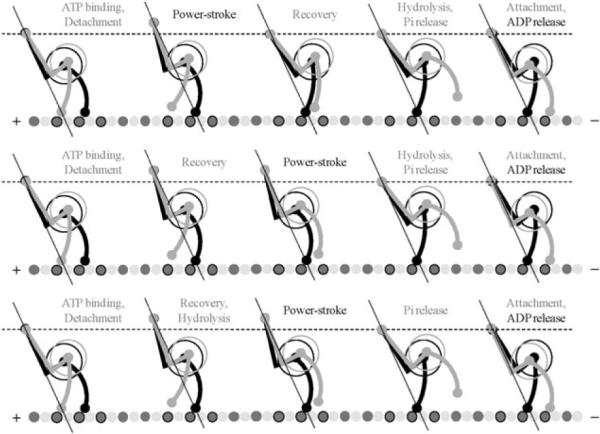
Detailed model for the two-headed dynein stepping consistent with both the two-headed kinetic model shown in Fig. 5 and experimentally observed conformational changes associated with pre-stroke and post-stroke states (Burgess et al.)1 Each of three stepping sequences corresponds to one of the parallel pathways in the kinetic model. The straight lines coming through the tail junction and one of the MTBD indicate the force line, along which the power-stroke opposes the external load. The load switches its carrier upon the detachment of the tailing head. The intermediate motion of the tail junction has both horizontal and vertical components, but each full cycle results in only horizontal displacement toward the minus end of the MT.
Upon ATP binding to the trailing head, this head loses its strong association with the microtubule and detaches. This leads to the load being transmitted to the leading head, so that the tail junction moves forward by ~8 nm and up by ~5 nm, which is the difference in mean length of dynein heads in post- and pre-stroke conformations. Then, the power stroke of the attached head moves tail junction down toward the microtubule by the same ~5 nm, along the force line. Now, the natural position for the head B to bind (after recovery, hydrolysis, and Pi release) will be in front of the head A to form the same two-headed conformation from which the cycle was started but with the reversed order of the heads. Finally, the release of ADP from the rear head completes the cycle.
It is important to notice that this picture is consistent with both Carter's model, which suggests force generation is in the direction toward the microtubule and Burgess's model, in which the power-stroke produces a change in the tail-stalk angle. Because we ignored flexibility of the tail and the stalk, the angle at which the stalk bound to the microtubule was determined by the geometry of the two heads. Thus, our next step is to consider both the tail and the stalk as an elastic object with fixed length but a variable curvature (see Fig. 7 and Appendix C100), which gives the best approximation to the shapes observed by Burgess et al.1
Fig. 7.
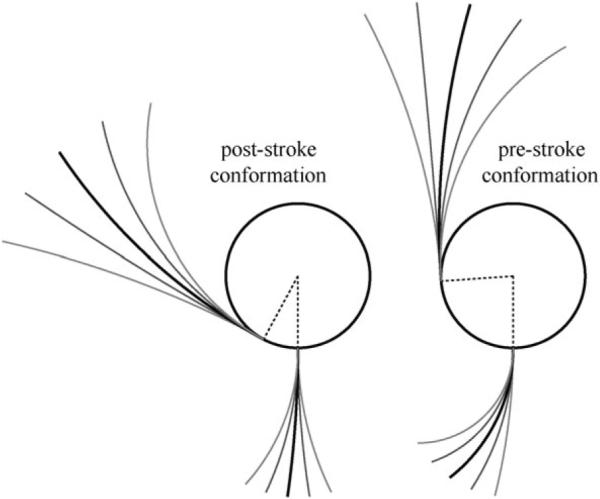
Shapes of dynein motor domains in post-stroke (left) and pre-stroke (right) states approximated by a circle (for AAA+ ring) and two arcs (for the stalk and the tail). The stalk and the tail emerge perpendicular and tangentially to the ring, respectively, while the curvature of the arcs is variable. The geometry was chosen to provide the best fit to shapes observed by Burgess et al.1 The range of flexibility and the mean (relaxed) positions of the stalk and the tail were defined to match the experimental observations. These parameters depend on the conformational state of motor unit.
5.2 Minimum energy conformation of the dimer
We choose a cost function associated for deviations from relaxed shape such that one standard deviation contributes one unit to the bending energy (see Appendix D500). Finally, we assume that the stalk binds to the microtubule at a right angle in both the pre-stroke and post-stroke states.
The kink that is associated with highly conserved prolines in the stalk is presumably responsible for the stalk's overall angled orientation with respect to the microtubule axis (leaning toward the plus end of the microtubule).35 This kinked conformation is accounted for in our simplified geometrical representation of the stalk.
One experiment that supports the attachment of the stalk to a MT at a fixed angle regardless of the nucleotide state was done using outer-arm dynein molecules purified from sea urchin sperm flagella.49 In this experiment, the authors imaged both dynein heads prepared in either pre-stroke or post-stroke states, and observed that the stalk orientation remained fixed with respect to the microtubule. However, the model derived by the authors is not necessarily applicable to cytoplasmic dynein. First, their model corresponds to inch-worm stepping rather than the head-over-head motion established for cytoplasmic dynein.11,13,50,51 Secondly, this model allows both heads to be in the weakly bound state, which again would be very unlikely for highly processive cytoplasmic dynein. On the other hand, the proposed mechanism of force generation, which involves retracting the elongated tail linker toward the AAA+ ring, is consistent with the observation that flipped-ring constructs maintain minus-end directed stepping,35 but only if the linker in the pre-stroke conformation is flexible enough and not specifically docked to the ring. This kind of “undocked” conformation was observed by Burgess et al.,1 but could not be identified as a pre-stroke or a post-stroke state. The uncertainty associated with the experimental results of Ueno et al. may be attributed to the difficulty with the preparation of bound pre-stroke heads and their flexibility, which could lead to the high variability seen in the ADP·Vi images. Thus, further experiments are needed to understand the nucleotide-dependent interaction between dynein's ring and linker.
We observed that under these assumptions the two head conformation, which require virtually no bending energy (e.g. both heads are in their relaxed shape), is when the leading and the trailing heads are attached 8.2 nm apart in the pre-stroke and post-stroke states, respectively (Fig. 8A and B).
Fig. 8.
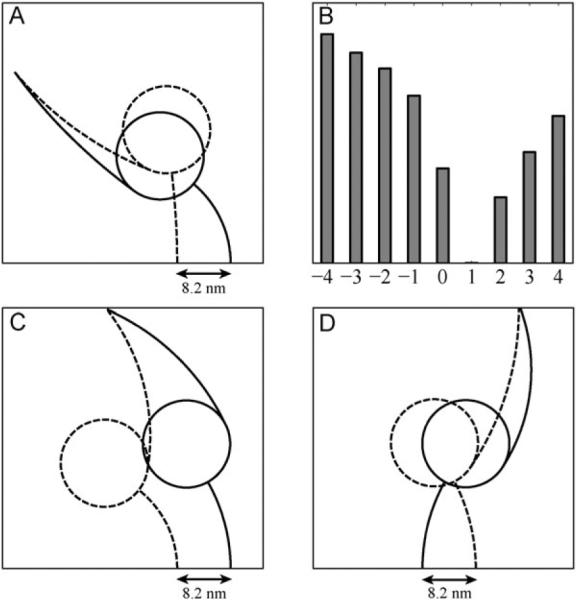
(A) The minimal bending energy geometry of dynein motor units in post-stroke (dashed lines) and pre-stroke (solid lines) states with their MTBD separated by 8.2 nm. At the points of attachment the stalk are enforced to be perpendicular to the MT; (B) Relative minimum bending energy (in arbitrary units) of the two-headed dynein as a function of the MTBD separation on the MT (in units of 8.2 nm). Plus and minus signs correspond to the attachment of the pre-stroke head in front and behind the other head, respectively. Here and throughout the paper `front' means toward the minus end of the MT; (C, D) The same as (A) but for the (−7) construct, in which the ring-tail complex of each head is flipped with respect to the stalk. All other geometrical parameters and conditions were left unaltered. The difference between (C) and (D) is in the relative position of the head's MTBDs on the microtubule, leading to the motion in the opposite directions according to the detailed two-headed model shown in Fig. 6.
This surprisingly good agreement with our predictions from the kinetic model motivates us to explore possible minimum energy two-head conformations of the (−7) construct in which seven heptads have been removed from the stalk,35 so that plains of the AAA+ rings are flipped by 180 degrees with respect to the microtubule binding domains.
5.3 Dynein stepping with flipped AAA+ ring
As demonstrated schematically in Fig. 9, if one accepts as a fact that the direction of stepping is defined by the attachment of the pre-stroke head with respect to the bound post-stroke head, then depending on their relative position flipped heads can adopt two-head conformations that move in the opposite directions. Thus, the single assumption that force generated by the power-stroke directed toward the microtubule along the stalk is not sufficient to guaranty that the two-headed (−7) construct steps in the same direction as the native motor. In other words, our two-headed kinetic model and simple geometric considerations suggest that both MTBD orientation and the relative ring-ring positioning are responsible for the direction of the motor's stepping.
Fig. 9.
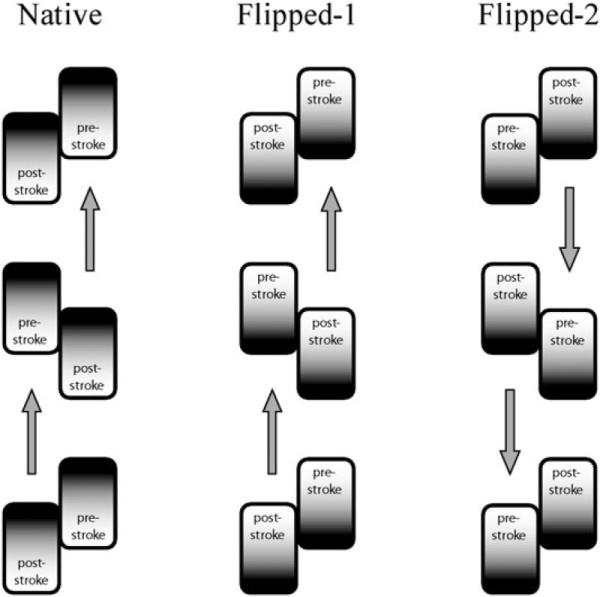
Schematic diagram demonstrating two possible two-headed orientations of the (−7) dynein construct with flipped AAA+ rings. Flipped-1 orientation maintains the relative to MT order of the pre-stroke and post-stroke states and, hence, maintains the direction of the stepping forward the minus end of the microtubule. Flipped-2 orientation corresponds to the flipping of the two-headed motor as a whole around the center of mass, and results in the reversed direction of stepping. For these considerations the direction of force generated by the power-stroke is irrelevant, as long as detached tailing head is biased to attach in the leading position and the heads are not in identical conformational states while they are attached.
With this in mind, we considered the minimum energy conformations for the two flipped orientations (Fig. 8C–D): when the post-stroke head is in front and behind the pre-stroke head with respect to minus end of the MT. We found that the orientation, which leads to plus-end directed motion, stores about twice as much bending energy as the minus-end stepping conformation. Hence, it is more likely that the flipped heads will maintain stepping direction of the native dynein, in agreement with the experiment.35 However, we should emphasize that for these calculations we have totally ignored the energy of the compact ring-ring conformation. If the formation of overlapping rings (docking) is much more energetically favorable than the specially separated rings, then the overall energy balance could be in favour of the plus-end directed (−7) construct. Otherwise, the non-compact orientation of the heads should disrupt their coordination and reduce the velocity of the stepping, again in agreement with the experimental observation.
We also found that the flipped position, in which the MTBD of the pre-stroke head is located more than 8.2 nm in front of the other head's MTBD, has even smaller bending energy. This observation suggests that the (−7) construct should take predominantly larger than 8.2 nm steps. However, we notice that in the above analysis we assumed that the (−7) construct has the same parameters for the stalk flexibility as the native head, which might not be the case. To confirm this prediction, the step-size distribution of the construct is needed. In any case, further development of the detailed stepping model for dynein requires both numerical and experimental studies of the interaction and mutual orientation of the AAA+ rings.
6. Outlook
Underlying the behaviour of molecular motors are motions that span multiple length and time scales. Thus, a detailed understanding of how they function requires a multi-scale approach. Such approaches have been successful in integrating atomic-level details of the mechano-chemical cycles and the experimentally single-molecule behaviours of F1F0 sythase,52–55 kinesin,56–59 and myosin.60,61 While such a description for dynein is lacking, the rapid increase in recent years in structural and single-molecule experimental investigations of this molecule are making a multi-scale modelling feasible. To investigate dynein's energy transduction mechanism of the atomic scale, we first constructed an atomic theoretical model of the motor unit, then subsequently performed molecular dynamics simulation. These simulations showed that the hydrolysis competent domains of the motor unit are less dynamic than the non-hydrolysing half. This observation suggests that the minor conformation induced by either hydrolysis or product release may be amplified by the more dynamic regions. On a much longer and larger time scale, we constructed kinetic models of the coordination between the two heads of a dynein dimer. From these models, we showed the various scenarios of coordination that satisfy the experimental observation of dynein's high processivity and high velocity. We likewise explore the potential geometries of the dimer during its chemomechanical cycle. Considering that dynein is structurally and evolutionary distinct from either F1F0 synthase, kinesin or myosin, both experimental and theoretical studies of this motor are likely to reveal novel mechanisms of energy transduction.
Fig. 5.
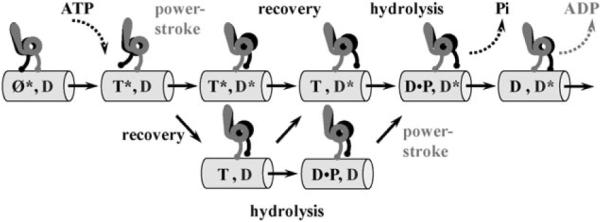
Schematic representation of the two-headed kinetic model for the dynein stepping.37 Grey and black labels for states and transitions correspond to the grey and black heads, respectively. Each head can be in one of four biochemical states: nucleotide-free (Ø), ATP-bound (T), ADP·Pi-bound state (D·P), and ADP-bound state (D), and simultaneously in one of two conformational states: post-stroke (*) and pre-stroke. Shown is only a subset of states, which corresponds to the dominant pathways in the full multi-state network.
Acknowledgements
We thank Dr Feng Ding for invaluable discussions on discrete molecular dynamics. Our work is supported by National Institutes of Health (R01-GM080742 to NVD) and (R01-GM079271 and R01-HL077546 to TCE). AWRS is supported by a pre-doctoral fellowship from the American Heart Association (0715215U).
Appendix A: simplified protein model
The protein model consisted only of Cα and Cβ atoms.26 The interaction potential used in the simulation is the structure-based Go-interaction,17,20 where the residues that were proximate in the native state were assigned an attractive interaction, but those that were not were assigned a repulsive interaction. The total potential energy of a model protein was then:
where i and j denoted residues i and j, Uij was the matrix of interactions:
Here, a0 was the hard core diameter, ai was the maximum interaction distance between residues and εij was the interaction strength between residue i and residue j, which set the energy scale. ∥Δij∥ was a matrix of contacts with elements:
where was the position of the ith residue in the native conformation. We penalized the non-native contacts by imposing εij < 0. Temperature units were taken in terms of the typical value of interaction strength εij divided by the Boltzmann constant kB, i.e., in units of εij/kB.
The strength of the interaction between residues in contact εij defines the energy units. Physically, εij ≈ 1–2 kcal mol−1, which is approximately the contribution to protein stability from a hydrogen bond. The time unit (tu) is estimated to be the shortest time between particle collisions in the system (~0.1 ns).
Appendix B: discrete molecular dynamics
The evolution of this simplified protein model with simplified atomic interactions was calculated using DMD. In contrast to traditional molecular dynamics which employs continuous potentials, the DMD algorithm uses discrete square well potentials,19,62–69 thus all particles move at constant velocity until and before the first collision. That the state of the system is necessarily updated only in the event of a collision enables DMD to access the long time scale dynamics of large proteins.
Appendix C: head geometry
We approximate the two-dimensional projection of each head shape as a disc of radius R and two circular arcs: one arc represents the stalk and emerges perpendicular to the disc perimeter, while the second arc is the tail and emerges tangentially from the disc (Fig. 10). The disc itself represents the AAA+ dynein ring. Each arc has a fixed length (Ls for the stalk and Lt for the tail) but a variable central angle (a for the stalk and β for the tail). Let us denote the tip of the microtubule domain as point A, the points of the stalk and the tail emergence from the disc as B and C, respectively, the tip of the tail as D and the center of the disc as O. Then the length of AB is equal to:
while the length of CD is equal to:
Finally, let as assume that the pre-stroke and post-stroke conformations of the head have different angles BOC, namely γpre and γpost, respectively. Now, if we define the coordinates of the point A, (xA,yB), and the angle at which the stalk is bound the horizontally positioned microtubule, δ, then the angles α and β fully determine the orientation of the head with the following coordinates of the points B, O, C and D:
The parameter values that we used in our simulations were extracted from the best fit to the experimental images by Burgess et al.1 and are given in the following Table 1:
Fig. 10.
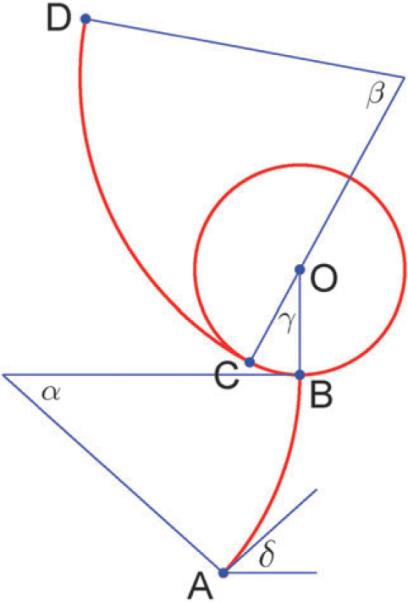
Head geometry illustration.
Appendix D: minimum energy geometry
Using the experimental images of Burgess et al.1 we determined the mean (equilibrium) position, mα and mβ, and the range of motion, σα and sb, of the stalk and the tail, respectively, for both pre-stroke and post-stroke conformations. Then, we define the relative bending energy for each head as:
To determine the minimum energy geometry of two-headed dynein we specify the head conformations, pre- or post-stroke, and the points of the stalk attachment to the microtubule, A1 and A2. Then we add a spring between tail tips, D1 and D2, with the zero value for the equilibrium length and a relatively
Table 1.
Parameter values for head geometry
| Ls/nm | Lt/nm | R/nm | γpre/° | γpost/° | δ/° |
|---|---|---|---|---|---|
| 13.9 | 26.0 | 6.7 | 86.0 | 28.5 | 90.0 |
Table 2.
Parameter values for minimum energy geometry of two-headed dynein
| Pre-stroke |
Post-stroke |
||||||
|---|---|---|---|---|---|---|---|
| mα/° | σα/° | mβ/° | σβ/° | mα/° | σα/° | mβ/° | σβ/° |
| 52 | 38 | 20 | 44 | 8 | 34 | 28 | 44 |
large spring constant, k, to ensure that the tail tips stay close enough to each other, and minimize the function:
where:
The angles at which F is minimal,
define the minimum energy two-headed geometry for the specified head attachments and their conformations, giving the total stored bending energy:
The parameter values that we used for our minimizations are given in the following Table 2:
Footnotes
Additional notes While the manuscript was under review, a paper describing a novel geometry of the dynein motor unit was published.70 The authors found that the core of the motor unit is composed of a hexamer of the AAA+ domains and that the C-domain spans between AAA6 and the base of the stalk. These findings run counter to some of our theories in section 2 and in ref. 2. We are currently in the process of revising the model to take these results into consideration. The issue of whether the proposed structure of the IDR4 is indeed coiled coil or not, and whether it spans the motor ring or not, is still a point of contention in the field. This issue may be resolved by higher resolution structural studies of the motor unit.
References
- 1.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 2.Serohijos AWR, Chen Y, Ding F, Elston TC, Dokholyan NV. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18540–18545. doi: 10.1073/pnas.0602867103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad FJ, Echeverri CJ, Vallee RB, Baas PW. J. Cell Biol. 1998;140:391–401. doi: 10.1083/jcb.140.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes JM, Katsanis N. Cell. Mol. Life Sci. 2005;62:1556–1570. doi: 10.1007/s00018-005-5007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuwald AF, Aravind L, Spouge JL, Koonin EV. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 6.Mocz G, Gibbons IR. Structure. 2001;9:93–103. doi: 10.1016/s0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 7.King SJ, Schroer TA. Nat. Cell. Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 8.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 9.Mallik R, Petrov D, Lex SA, King SJ, Gross SP. Curr. Biol. 2005;15:2075–2085. doi: 10.1016/j.cub.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Khan S, Sheetz MP. Biophys. J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 12.Vale RD, Milligan RA. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 13.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samso M, Koonce MP. J. Mol. Biol. 2004;340:1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 15.Lee SY, De la Torre A, Yan DL, Kustu S, Nixon BT, Wemmer DE. Genes Develop. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dokholyan NV. Curr. Opin. Struct. Biol. 2006;16:79–85. doi: 10.1016/j.sbi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Abe H, Go N. Biopolymers. 1981;20:1013–1031. doi: 10.1002/bip.1981.360200512. [DOI] [PubMed] [Google Scholar]
- 18.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Folding Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 19.Ding F, Dokholyan NV. Trends Biotechnol. 2005;23:450–455. doi: 10.1016/j.tibtech.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Go N, Abe H. Biopolymers. 1981;20:991–1011. doi: 10.1002/bip.1981.360200511. [DOI] [PubMed] [Google Scholar]
- 21.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. J. Mol. Biol. 2000;296:1183–1188. doi: 10.1006/jmbi.1999.3534. [DOI] [PubMed] [Google Scholar]
- 22.Dokholyan NV, Li L, Ding F, Shakhnovich EI. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8637–8641. doi: 10.1073/pnas.122076099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khare SD, Ding F, Dokholyan NV. J. Mol. Biol. 2003;334:515–525. doi: 10.1016/j.jmb.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 24.Chen YW, Dokholyan NV. J. Biol. Chem. 2006;281:29148–29154. doi: 10.1074/jbc.M605512200. [DOI] [PubMed] [Google Scholar]
- 25.Borreguero JM, Dokholyan NV, Buldyrev SV, Shakhnovich EI, Stanley HE. J. Mol. Biol. 2002;318:863–876. doi: 10.1016/S0022-2836(02)00136-5. [DOI] [PubMed] [Google Scholar]
- 26.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Biophys. J. 2002;83:3525–3532. doi: 10.1016/S0006-3495(02)75352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S, Ding F, Dokholyan NV. Biophys. J. 2007;92:1457–1470. doi: 10.1529/biophysj.106.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks B, Karplus M. Proc. Natl. Acad. Sci. U. S. A. 1983;80:6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahar I, Rader AJ. Curr. Opin. Struct. Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee MA, Heuser JE, Vallee RB. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 31.Gee MA, Heuser JE, Vallee RB. Mol. Biol. Cell. 1997;8:930–930. [Google Scholar]
- 32.Woehlke G, Ruby AK, Hart CL, Ly B, HomBooher N, Vale RD. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 33.Rayment I, Rypniewski WR, Schmidtbase K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. J. Biol. Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolomeisky AB, Fisher ME. Ann. Rev. Phys. Chem. 2007;58:675–695. doi: 10.1146/annurev.physchem.58.032806.104532. [DOI] [PubMed] [Google Scholar]
- 37.Miki H, Okada Y, Hirokawa N. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Hirokawa N, Noda Y. Phys. Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 39.Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. J. Struct. Biol. 2006;156:182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Shima T, Kon T, Imamula K, Ohkura R, Sutoh K. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17736–17740. doi: 10.1073/pnas.0606794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 42.Tsygankov D, Serohijos AWR, Dokholyan NV, Elston TC. J. Chem. Phys. 2009;130:025101–025113. doi: 10.1063/1.3050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reck-Peterson SL, Vale RD. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1491–1495. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Singh MP, Mallik R, Gross SP, Yu CC. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12059–12064. doi: 10.1073/pnas.0501570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho C, Reck-Peterson SL, Vale RD. J. Biol. Chem. 2008 doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 48.Tsygankov D, Elston TC. Biophys. J. 2009;96:507a. [Google Scholar]
- 49.Ueno H, Yasunaga T, Shingyoji C, Hirose K. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19702–19707. doi: 10.1073/pnas.0808194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oster G, Wang HY. Trends Cell Biol. 2003;13:114–121. doi: 10.1016/s0962-8924(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 53.Sun SX, Wang HY, Oster G. Biophys. J. 2004;86:1373–1384. doi: 10.1016/S0006-3495(04)74208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao YQ, Yang W, Karplus M. Cell. 2005;123:195–205. doi: 10.1016/j.cell.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Pu JZ, Karplus M. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1192–1197. doi: 10.1073/pnas.0708746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fisher ME, Kolomeisky AB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7748–7753. doi: 10.1073/pnas.141080498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolomeisky AB, Fisher ME. Physica A. 2000;279:1–20. [Google Scholar]
- 58.Hyeon C, Onuchic JN. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17382–17387. doi: 10.1073/pnas.0708828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyeon C, Onuchic JN. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui Q. Biophys. J. 2004;86:404A–404A. doi: 10.1016/S0006-3495(04)74152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischer S, Windshugel B, Horak D, Holmes KC, Smith JC. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding F, LaRocque JJ, Dokholyan NV. J. Biol. Chem. 2005;280:40235–40240. doi: 10.1074/jbc.M506372200. [DOI] [PubMed] [Google Scholar]
- 63.Chen YW, Dokholyan NV. J. Mol. Biol. 2005;354:473–482. doi: 10.1016/j.jmb.2005.09.075. [DOI] [PubMed] [Google Scholar]
- 64.Khare SD, Ding F, Gwanmesia KN, Dokholyan NV. Plos Comput. Biol. 2005;1:230–235. doi: 10.1371/journal.pcbi.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khare SD, Wilcox KC, Gong P, Dokholyan NV. Proteins Struct. Funct. Bioinfor. 2005;61:617–632. doi: 10.1002/prot.20629. [DOI] [PubMed] [Google Scholar]
- 66.Zhou YQ, Karplus M. Nature. 1999;401:400–403. doi: 10.1038/43937. [DOI] [PubMed] [Google Scholar]
- 67.Peng S, Ding F, Urbanc B, Buldyrev SV, Cruz L, Stanley HE, Dokholyan NV. Phys. Rev. E. 2004;69 doi: 10.1103/PhysRevE.69.041908. [DOI] [PubMed] [Google Scholar]
- 68.Ding F, Prutzman KC, Campbell SL, Dokholyan NV. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 69.Ding F, Jha RK, Dokholyan NV. Structure. 2005;13:1047–1054. doi: 10.1016/j.str.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 70.Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, Burgess SA. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]