Abstract
We report the isolation from tomato (Lycopersicon esculentum) of an ethylene-responsive member of the proteinase inhibitor gene family. DNA sequence analysis of a full-length cDNA clone indicates that the ethylene-responsive gene is distantly related to the tomato proteinase inhibitor I gene, having 53% sequence identity. The predicted amino acid sequence reveals 47% and 45% sequence identity with the tomato and potato proteinase inhibitor I polypeptides, respectively. Additionally, the ethylene-responsive inhibitor has evolved a completely different pattern of gene expression and inhibitory specificity than other members of the inhibitor I family. Gel blot hybridization experiments show that, unlike the tomato proteinase inhibitor I gene, it is not induced in wounded leaves. In contrast, it is activated by the plant hormone ethylene in leaves and during fruit ripening. Furthermore, the ethylene-responsive inhibitor exhibits a novel reactive site, having glutamic acid as the P1 residue. This suggests that the ethylene-responsive proteinase inhibitor does not react with chymotrypsin, as does proteinase inhibitor I, but that it reacts with proteolytic enzymes that cleave at glutamic residues, such as the Staphylococcus aureus V8 proteinase, for which no inhibitors are known. Finally, isolation and analysis of a genomic clone reveals that the ethylene-responsive proteinase inhibitor gene is tightly linked to another, yet unidentified, coordinately expressed gene. We discuss these results with regard to the function and evolution of proteinase inhibitor genes in tomato.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Dellapenna D., Alexander D. C., Bennett A. B. Molecular cloning of tomato fruit polygalacturonase: Analysis of polygalacturonase mRNA levels during ripening. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R. L., Goldberg R. B. Structure and flanking regions of soybean seed protein genes. Cell. 1982 Jun;29(2):651–660. doi: 10.1016/0092-8674(82)90181-7. [DOI] [PubMed] [Google Scholar]
- Graham J. S., Pearce G., Merryweather J., Titani K., Ericsson L. H., Ryan C. A. Wound-induced proteinase inhibitors from tomato leaves. II. The cDNA-deduced primary structure of pre-inhibitor II. J Biol Chem. 1985 Jun 10;260(11):6561–6564. [PubMed] [Google Scholar]
- Graham J. S., Pearce G., Merryweather J., Titani K., Ericsson L., Ryan C. A. Wound-induced proteinase inhibitors from tomato leaves. I. The cDNA-deduced primary structure of pre-inhibitor I and its post-translational processing. J Biol Chem. 1985 Jun 10;260(11):6555–6560. [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hill R. E., Hastie N. D. Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature. 1987 Mar 5;326(6108):96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- Hill R. E., Shaw P. H., Boyd P. A., Baumann H., Hastie N. D. Plasma protease inhibitors in mouse and man: divergence within the reactive centre regions. Nature. 1984 Sep 13;311(5982):175–177. doi: 10.1038/311175a0. [DOI] [PubMed] [Google Scholar]
- Houmard J., Drapeau G. R. Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T. W., Storms S., Young P., Phillips L. E., Rogers T. E., Moore D. G., Williams R. P. Reactivity of microhemagglutination, fluorescent treponemal antibody absorption, Venereal Disease Research Laboratory, and rapid plasma reagin tests in primary syphilis. J Clin Microbiol. 1983 Mar;17(3):405–409. doi: 10.1128/jcm.17.3.405-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I., Ardelt W., Cook J., Denton A., Empie M. W., Kohr W. J., Park S. J., Parks K., Schatzley B. L. Ovomucoid third domains from 100 avian species: isolation, sequences, and hypervariability of enzyme-inhibitor contact residues. Biochemistry. 1987 Jan 13;26(1):202–221. doi: 10.1021/bi00375a028. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Brown W. E., Graham J. S., Pearce G., Fox E. A., Dreher T. W., Ahern K. G., Pearson G. D., Ryan C. A. Molecular characterization and phylogenetic studies of a wound-inducible proteinase inhibitor I gene in Lycopersicon species. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7277–7281. doi: 10.1073/pnas.83.19.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
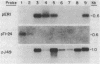
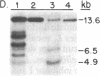
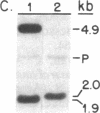
- Lincoln J. E., Cordes S., Read E., Fischer R. L. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc Natl Acad Sci U S A. 1987 May;84(9):2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Richardson M., Cossins L. Chymotryptic inhibitor I from potatoes: the amino acid sequences of subunits B, C, and D. FEBS Lett. 1974 Sep 1;45(1):11–13. doi: 10.1016/0014-5793(74)80798-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]