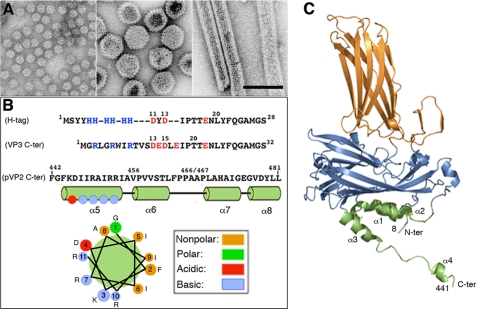
FIGURE 1.
(p)VP2-based chimeric protein system to test VP2 structural polymorphism. A, left, major (p)VP2 regular assemblies isolated from IBDV-infected avian cells or, using heterologous expression systems, from mammalian cells with an inducible rVV and in insect cells with the baculovirus expression system. Icosahedral T = 1 subviral particles (∼23-nm diameter) are built by VP2 trimers associated in an all-pentamer capsid. Center, T = 13 virion particles (65–70-nm diameter) are formed by 260 VP2 trimers making 12 pentamers and 120 hexamers. Right, tubular structures (45–60-nm diameter, depending on the number of helical rows) with a hexagonal lattice are all-hexamer structures. B, VP3 and pVP2 polypeptide segments involved in VP2 structural polymorphism are shown. His tag (H-tag) and VP3 C-terminal (VP3 C-ter) sequences are aligned based on polarity. The His tag used here is composed of a tract of four residues (Met1-Tyr4), six His residues, a linker region (segment Asp11-Thr17), the TEV cleavage site (Glu18-Gly24), and the tract Asn25-Ser28, to which the (p)VP2-based protein is bound. The VP3 C terminus includes the last 16 VP3 residues (242GRLGRWIRTVSDEDLE257; for simplicity, Glu257 is referred to here as Glu17, and so on) and the linker region (except the sequence 11DYD13), the TEV cleavage site (Glu18-Gly24), and the tract Asn25-Ser28. The C-terminal (p)VP2 Phe442-Leu481 sequence (pVP2 C-ter) is shown. The amphipathic α-helix (residues 443–452) is referred as helix α5; α6-, α7-, and α8-helices were also determined by NMR analysis (16). A helical wheel representation of the amphipathic α-helix is shown (bottom). C, VP2 protein x-ray model (Protein Data Bank code 2GSY). Domains P, S, and B are colored orange, blue, and green, respectively. The α-helices (1–4) of domain B are indicated. Note that N and C termini are relatively close.