Abstract
The strychnine-sensitive glycine receptor (GlyR) is a ligand-gated ion channel that mediates fast synaptic inhibition in the vertebrate central nervous system. As a member of the family of Cys-loop receptors, it assembles from five homologous subunits (GlyRα1–4 and -β). Each subunit contains an extracellular ligand binding domain, four transmembrane domains (TM), and an intracellular domain, formed by the loop connecting TM3 and TM4 (TM3–4 loop). The TM3–4 loops of the subunits GlyRα1 and -α3 harbor a conserved basic motif, which is part of a potential nuclear localization signal. When tested for functionality by live cell imaging of green fluorescent protein and β-galactosidase-tagged domain constructs, the TM3–4 loops of GlyRα1 and -α3, but not of GlyRα2 and -β, exhibited nuclear sorting activity. Subunit specificity may be attributed to slight amino acid alterations in the basic motif. In yeast two-hybrid screening and GST pulldown assays, karyopherin α3 and α4 were found to interact with the TM3–4 loop, providing a molecular mechanism for the observed intracellular trafficking. These results indicate that the multifunctional basic motif of the TM3–4 loop is capable of mediating a karyopherin-dependent intracellular sorting of full-length GlyRs.
Keywords: Nucleus/Nuclear Import, Protein/Sorting, Intracellular Trafficking, Membrane Proteins, Receptor Structure-Function, Glycine Receptor, Karyopherin, Multifunctional Motif, Sorting, Trafficking
Introduction
In the vertebrate nervous system, signal transmission at chemical synapses is mediated by ionotropic and metabotropic neurotransmitter receptors. Ligand-gated ion channels harbor an intrinsic channel pore that is opened almost instantly upon ligand binding. Among the ligand-gated ion channels, the superfamily of Cys-loop receptors comprises the nicotinic acetylcholine receptor, the 5-hydroxytryptamine type 3 receptor, the γ-aminobutyric acid type A/C receptor, and the GlyR.2 Besides sequence homology, Cys-loop receptors share a common pentameric rosette-like composition of homologous subunits and a characteristic subunit topology (Fig. 1, A and B). The N-terminal extracellular domain forms a twisted β-sandwich structure contributing to the intersubunit ligand binding pockets (1). Each subunit contains four α-helical TMs, where TM2 lines the channel pore. The intracellular domain of the receptor is mainly formed by the TM3–4 loop; its protein conformation has not been resolved. The C-terminal part of the TM3–4 loop, however, is thought to form an intracellular cavity that might contribute to ion selectivity (2).
FIGURE 1.
Structural levels of the GlyR. A, a schematic representation is shown of the GlyR as a pentameric rosette-like complex of homologous subunits (ovals) embedded in the membrane. B, putative topology of the glycine receptor subunits based on the structure of the nicotinic acetylcholine receptor is shown (1, 2). β-Strands are depicted as arrows, and α-helices are depicted as barrels. C, shown is multiple polypeptide sequence alignment of the TM3–4 loops of human (H. sapiens, hs), rat (R. norvegicus, rn), and mouse (M. musculus, mm) GlyR subunits as annotated in the Uniprot data base including amino acid positions within the respective precursors. Known splice variants resulting in variations within the TM3–4 loop are included. The alignment is arbitrarily subdivided into conserved motifs (C1–3) and variable regions (V1 and V2). The homology between α subunits as well as the overall homology is indicated. The sequences of human subunits aligned with the basic motif 346RRKRR350 of hsGlyRα1 are underlined.
Spinal GlyRs play an important role in the neuronal control of muscle tone. Upon glycine binding, the intrinsic chloride channel opens, resulting in hyperpolarization of the postsynaptic membrane of α motoneurons, thereby reducing their activity. GlyR defects underlay excessive reflex responses, as in symptomatic hyperekplexia (OMIM 149400), stiff-person syndrome (OMIM 184850), or strychnine intoxication (3). Up to five GlyR genes have been described in vertebrates (4), four of which code for subunits that are active as homomers in heterologous systems (α1–4). Among these, the α2 subunit predominates during embryonic development, forming homomeric, extrasynaptic receptors that are thought to be important for calcium-dependent synaptogenesis (5, 6). The adult receptors, however, are mostly heteromers of 2 GlyRα and 3 GlyRβ subunits, both of which contribute to ligand binding (7).
The TM3–4 loop is the domain with the highest sequence variability between different subunits, possibly regulating trafficking, anchoring, and protein interactions. The best characterized interaction takes place between gephyrin and a 13-residue motif in the TM3–4 loop of GlyRβ (8). Gephyrin forms hexagonal lattices at the postsynaptic density, allowing for synaptic anchoring of heteromeric receptors (9, 10). As a postsynaptic scaffold for glycinergic synapses, gephyrin also recruits a number of interaction partners to the synapse (11). By interaction with the dynein complex, gephyrin finally mediates vesicular retrograde transport of the receptor along microtubules (12). For the TM3–4 loops of GlyRα subunits, a number of protein-protein interactions that are guided by consensus motifs have been described (13, 14). However, the roles of these interactions and consensus motifs in trafficking of the receptor remain elusive.
Here, we report the presence of a multifunctional basic motif that is part of a potential nuclear localization signal (NLS) within the TM3–4 loop of adult, synaptic GlyRα subunits, i.e. GlyRα1 and -α3. When functionality of this NLS was tested, full-length GlyR subunits, but not fragments, were shown to be imported into the nucleus. Karyopherins α3 and α4 were identified as interaction partners of the GlyR in vitro. In addition to nuclear sorting, this interaction may also underlie other functions of the basic motif, including preservation of correct protein topology or membrane integration.
EXPERIMENTAL PROCEDURES
Sequence Analysis and Cloning
For multiple sequence alignment, the sequences and boundaries of the TM3–4 loops were taken from the annotation in the Uniprot data base (15). Alternative variants of the TM3–4 loops of human (Homo sapiens, hs), rat (Rattus norvegicus, rn), and mouse (Mus musculus, mm) GlyR subunits were subjected to a multiple sequence alignment using the T-COFFEE algorithm (16). Prediction of subcellular localization and nuclear import was performed with the TM3–4 loop sequence of hsGlyRα1. Cloning was carried out with standard protocols using plasmid DNA as template and overlap extension PCR for mutagenesis. Fusion proteins with green fluorescent protein (GFP) were generated using the pEGFP system (N1, C1–3; Clontech, Mountain View, CA). Because of high sequence conservation, orthologous GlyR subunits were used equivalently. The cloned TM3–4 loops of human and murine GlyR subunits were identical to the sequences obtained from the Uniprot data base, whereas for historical reasons the TM3–4 loops of rat GlyRα subunits include the additional N-terminal sequence of AAVNFVS and the TM3–4 loop of rat GlyRβ carries the additional N-terminal sequence of VVQVML, which are annotated as TM3 in the Uniprot data base. For enlargement of the final fusion constructs, β-galactosidase full-length cDNA was inserted in-frame into pEGFP-N1 making use of NheI and BglII restriction sites to preserve the multiple cloning site for insertion of the TM3–4 loop. Karyopherin full-length cDNAs were obtained from imaGenes, Berlin. For GST pulldown assays, the cloning vectors pET21a and pET41a were used (EMD, La Jolla, CA).
Yeast Two-hybrid Screening and GST Pulldown Assay
8,000,000 cDNAs from an adult human brain Matchmaker cDNA library in pACT2 (Clontech; HL4004AH) were screened with the TM3–4 loop of rnGlyRα3L as bait. In specificity tests, interaction of single clones with other GlyR subunit TM3–4 loops was tested. A GST pulldown assay was performed essentially as described (17).
Neuronal Cell Culture
Hippocampi were isolated from Wistar rats at embryonal day 18, meninges were removed, and tissue was pooled. Trypsin digestion was carried out with 1 mg/ml trypsin, 0.1 mg/ml DNase I in neurobasal medium (Invitrogen) at 37 °C for 30 min and stopped by the addition of 10% (v/v) fetal calf serum. Cells were dissociated by trituration, washed, and plated in neurobasal medium with 1% B-27 (Invitrogen) in 3-cm cell culture dishes or in 24-well plates on polylysine-coated coverslips. Transfection was carried out using Lipofectamine (Invitrogen) according to the manufacturer's instructions.
Antibodies for Immunocytochemistry, Immunoprecipitation, and Western Blot
Primary antibodies were mouse-anti-GlyRα (mAb4a, purified), mouse-anti-GlyRα1 (mAb2b purified, kindly provided by Dr. Heinrich Betz, MPI, Frankfurt, Germany), rabbit-anti-lamin A and mouse-anti-hemagglutinin (HA) tag (Abcam, Cambridge, UK), goat-anti-karyopherin α3 and goat-anti-Karyopherin α4 (Everest, Upper Heyford, UK), rabbit-anti-synaptophysin (Millipore, Billerica, MA), rabbit-anti-neurofilament M (Millipore), mouse-anti-GFP (Clontech), rabbit-anti- glyceraldehyde-3-phosphate dehydrogenase and rabbit-anti-histone deacetylase 1 (Santa Cruz Biotechnology, Santa Cruz, CA), and mouse-anti-T7-tag (EMD). Secondary antibodies were donkey-anti-goat-Alexa 488 (Invitrogen), donkey-anti-rabbit-Cy5, donkey-anti-mouse-Cy3 for immunocytochemistry, and goat-anti-mouse horseradish peroxidase (HRP) coupled as well as goat-anti-rabbit HRP for Western blot (Dianova, Hamburg, Germany).
Immunocytochemistry, GFP Imaging, and Electrophysiology
For immunocytochemistry, HEK293 cells or neurons were grown on polylysine-coated coverslips, transfected if necessary, fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS), and permeabilized as well as blocked with 0.1% Triton X-100, 5% donkey or sheep serum (Dianova) in PBS for 30 min at room temperature. Primary and secondary antibodies were diluted 1:200–400 in 5% serum. Cells were incubated with primary antibodies for 1 h at room temperature and washed 3 times with PBS. This procedure was repeated with secondary antibodies. Finally, cells were embedded in Mowiol and subjected to confocal microscopy on a DMIRE2 confocal microscope (Leica, Wetzlar. Germany). If required, serial z-scans were performed. Medial z-slices were used to define central nucleoplasmic regions of interest analyzed for fluorescence intensity using Leica Confocal Software. For GFP-imaging, HEK293 cells were grown on chamber slides (ibidi, Martinsried, Germany) and transfected by calcium phosphate precipitation. After 12–36 h, medium was exchanged for PBS followed by live cell imaging. Alternatively, transfected neurons grown on polylysine-coated coverslips were fixed using 5% acetate in methanol for 10 min at −20 °C, embedded in Mowiol, and subjected to either confocal or standard fluorescence microscopy, the latter with an Axioskop microscope (Carl Zeiss, Jena, Germany). Electrophysiology was carried out essentially as described (18).
Immunoprecipitation and Western Blot
For immunoprecipitation, either cellular compartments were used or whole-cell lysates were prepared. The latter was accomplished by homogenization of tissue or cultured cells in 50 mm Tris, 150 mm NaCl, 2% (v/v) Triton X-100, 5 mm EDTA, and complete protease inhibitors (Hoffmann-La Roche), pH 7.4, and clearance by centrifugation for 1 h at 100,000 × g and 4 °C. To the respective solution, antibodies with 4–10 μg/ml final concentration were added, mixed, and agitated overnight at 4 °C. 20 μl of Pansorbin-Agarose (EMD) was added, and agitation on 4 °C was continued for 2 h. Then, the beads were washed four times with PBS before they were subjected to Western blot analysis. Primary antibodies were diluted 1:2000, and secondary horseradish peroxidase-coupled antibodies were diluted 1:20000. Chemiluminescence substrates were ECL+ (GE Healthcare) and SuperSignal West Femto (Thermo Fischer Scientific, Waltham, MA) for normal and weak signals, respectively.
Cellular Fractionation Techniques
Multiple cell fractions were obtained with Compartmental Protein Extraction kit according to the manufacturer's instructions (Millipore). Alternatively, cells were grown on a 10-cm cell culture dish, washed twice in PBS, harvested in 400 μl of Buffer A (10 mm HEPES, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 2 mm dithiothreitol, and complete protease inhibitors (Hoffmann-La Roche), pH 7.9) and incubated on ice for 5 min. By the addition of 1% (final concentration) Nonidet P-40 and mixing, cells were lysed. The supernatant after centrifugation for 30 s at 20,000 × g and 4 °C was taken as the extranuclear compartment. Subsequently, the pellet was resuspended in 200 μl of Buffer B (Buffer A including 400 mm NaCl), mixed, and agitated at 4 °C for 15 min and centrifuged for 5 min at 20,000 × g and 4 °C. The supernatant was taken as the nuclear compartment. Membrane preparations were performed essentially as described (18). For alkaline extraction, 100 μl of membrane preparation was centrifuged at 100,000 × g and 4 °C for 1 h. The pellet was resuspended in 100 μl of 100 mm Na2CO3, pH 11.5, by sonification with a microtip (15 × 3 s on ice), incubated on ice for 30 min, and centrifuged at 100,000 × g and 4 °C for 1 h. The supernatant (corresponding to the membrane associated proteins) was saved, and the pellet (corresponding to integral membrane proteins) was resuspended in 100 μl of PBS.
RESULTS
Sorting Prediction and Sequence Analysis
To predict potential sorting motifs within the TM3–4 loop of human GlyRα1 (hsGlyRα1), we used different algorithms (Table 1). Curiously, both PSORTII (19) and HSLPred (20) predicted the nucleus to be the most probable sorting destination of the TM3–4 loop. Next, we applied two algorithms specialized for nuclear proteins. PredictNLS (21), which is restricted to published NLSs in a given polypeptide sequence, did not provide a significant hit. NucPRED, however, confirmed the nuclear sorting with considerable specificity and identified the basic motif 346RRKRR350 as a potential NLS (22). By multiple sequence alignment, we determined that this motif was highly conserved in GlyRα but not β subunits (Fig. 1C).
TABLE 1.
Prediction of sorting signals
| Algorithm | Result | Confidence |
|---|---|---|
| PSORT II | Nuclear localization | 65.2% |
| HSLPred | Nuclear localization | Reliability index 1, expected accuracy 51.02% |
| NucPRED | Nuclear localization predicted NLS motif: RRKRR | Specificity > 81% |
| PredictNLS | Sequence contains no known NLS | Not indicated |
Nuclear Accumulation of the TM3–4 Loop
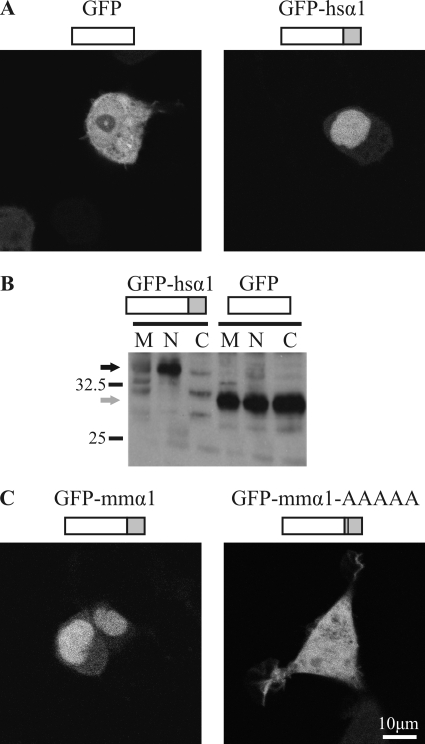
To determine the actual subcellular localization of the GlyRα1 TM3–4 loop, we generated fusion constructs (Fig. 2) with GFP and performed live cell imaging of HEK293 cells transfected with these plasmids (Fig. 2, A and C). As expected, GFP distributed evenly between cytoplasm and nucleus. By contrast, fusion constructs of GFP with the TM3–4 loops from human or mouse GlyRα1 accumulated in the nucleus. This is consistent with the high sequence similarity between species orthologs seen in multiple sequence alignments (Fig. 1C). Nuclear accumulation was confirmed, applying cell fractionation by differential centrifugation and Western blot using an antibody against GFP (Fig. 2B). When the basic motif (RRKRR) was mutated to a polyalanine sequence in the mouse GlyRα1 TM3–4 loop construct, the resulting fusion protein distributed evenly between nucleus and cytoplasm, as observed for GFP (Fig. 2B). This indicated that the TM3–4 loop holds a five-amino acid basic sorting motif that is necessary for the promotion of nuclear accumulation.
FIGURE 2.
Intracellular distribution of the TM3–4 loop. A, shown is confocal live microscopy of HEK293 cells transfected with GFP and GFP C-terminal fused to the TM3–4 loop of human GlyRα1 (GFP-hsα1). Representative examples are shown. B, shown is a Western blot of cellular fractions of HEK293 cells transfected with GFP-hsα1 (black arrow) or GFP (gray arrow). Fractions are membrane (M), nucleus (N), and cytoplasm (C). Staining was performed with an anti-GFP-antibody. C, confocal live microscopy of HEK293 cells transfected with C-terminal fusion constructs of the TM3–4 loop of murine GlyRα1 (GFP-mmα1) and mmGlyRα1 with the motif 346RRKRR350 replaced by five alanines (GFP-mmα1-AAAAA). Representative examples are shown. The TM3–4 loops of GlyRα1 of different species are accumulated in the nucleus, and the basic motif is necessary for this targeting.
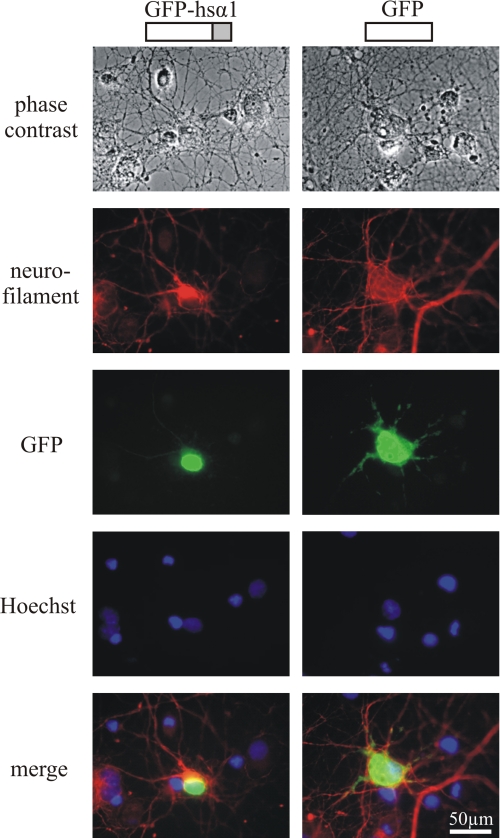
To test whether the TM3–4 loop containing the motif RRKRR was also active as a sorting signal in nerve cells, we transfected primary rat hippocampal neurons with these constructs (Fig. 3). As evident from nuclear accumulation, the cellular activity of the basic sorting motif in neurons was similar to that of HEK293 cells. Apparently, HEK293 cells provide an appropriate model to study this sorting mechanism.
FIGURE 3.
Distribution of the GlyRα1 TM3–4 loop in neurons. Shown is fluorescence microscopy of hippocampal neurons transfected with GFP or a fusion protein of GFP and the TM3–4 loop of human GlyRα1 (hsα1-GFP). For comparison, phase contrast, nuclear staining (Hoechst), and anti-neurofilament staining are provided. Representative examples are shown. Nuclear accumulation of the GlyRα1 TM3–4 loop also occurs in neuronal cells.
Only Selected GlyR Subtypes Harbor a Functional NLS
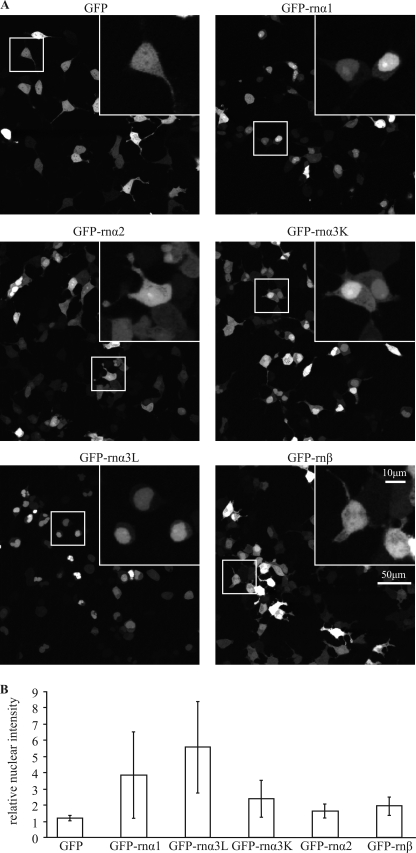
To test the impact of sequence diversity between different GlyR subunits on nuclear accumulation, we transfected HEK293 cells with GFP and GFP C-terminal-fused to TM3–4 loops of rnGlyRα1, rnGlyRα2, rnGlyRα3K, rnGlyRα3L, or rnGlyRβ. In live confocal microscopy, large intercellular variability of absolute and relative nuclear fluorescence intensity was observed for all TM3–4 loop fusion constructs. Nevertheless, calculation of nuclear to cytoplasmic fluorescence intensity ratios indicated pronounced nuclear accumulation for the TM3–4 loops of rnGlyRα1 and rnGlyRα3L and little nuclear accumulation for rnGlyRα3K (Fig. 4A). As expected from the multiple sequence alignment, which revealed little conservation of the required basic motif RRKRR (Fig. 1C), the TM3–4 loop of rnGlyRβ showed no tendency to nuclear accumulation. Surprisingly, the TM3–4 loop of GlyRα2 also failed to induce nuclear accumulation. This may be related to the interruption of the basic motif by a glutamine residue in the sequence of GlyRα2 (RRRQK).
FIGURE 4.
Subunit specificity of relative distribution. A, shown is confocal live microscopy of HEK293 cells transfected with GFP and GFP C-terminally fused to the TM3–4 loop of rat GlyRα1 (GFP-rnα1), GlyRα2 (GFP-rnα2), GlyRα3K (GFP-rnαK), GlyRα3L (GFP-rnα3L), and GlyRβ (GFP-rnβ). Representative examples are shown. Boxed segments are enlarged in the upper right panels. B, quantification of relative nuclear intensity with units representing -fold cytosolic fluorescence intensity is shown. GFP-rnα1 and GFP-rnα3L show strong nuclear accumulation, GFP-rnαK shows slight nuclear accumulation, and GFP-rnα2 and GFP-rnβ show no nuclear accumulation.
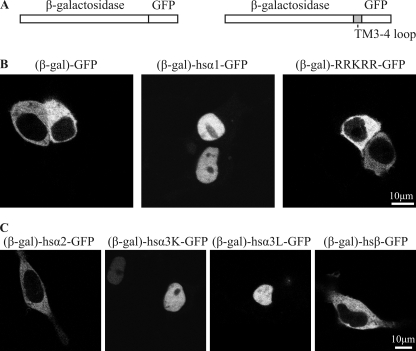
Subunit-specific Active Nuclear Import
Given their small size of <40 kDa, it cannot be excluded that nuclear accumulation of the recombinant fusion proteins resulted from diffusion through the nuclear pore and subsequent intranuclear retention. Diffusion through the nuclear pore, however, can be restricted by enlargement of the molecular weight of the fusion protein (23). To this end, we fused β-galactosidase to the reporter constructs, obtaining molecular masses >150 kDa (Fig. 5A). Although the β-galactosidase-GFP fusion was efficiently excluded from the nucleus, we observed nuclear import for fusion constructs of TM3–4 loop of GlyRα1 with β-galactosidase and GFP (Fig. 5B). However, a construct composed of merely the basic motif 346RRKRR350 fused between β-galactosidase and GFP with terminal spacer sequences was excluded from the nucleus. This suggests that the basic motif is required but not completely sufficient for nuclear import. Subsequently, we tested the TM3–4 loop sequences of different GlyR subunits including the long (L) and short (K) splice variants of GlyRα3 (24) for the presence of a functional NLS. As observed for GlyRα1, the TM3–4 loops of the GlyRα3L and α3K (potential NLS: RRKRK) also mediated nuclear import of β-galactosidase-GFP fusion proteins. By contrast, the TM3–4 loops of GlyRα2 and -β failed to show nuclear import (Fig. 5C) essentially confirming the results obtained with GFP-TM3–4-loop fusion constructs (Fig. 4). Thus, the TM3–4 loops of GlyRα1, -α3L, and -α3K subunits each contain a functional NLS, which accounts for the nuclear accumulation observed previously (Fig. 4).
FIGURE 5.
Subunit-specific active nuclear import of the TM3–4 loop. A, a schematic representation of the constructs used is shown. The fusion protein of GFP and β-galactosidase cannot pass the nuclear pore by diffusion due to its size (>150 kDa). Using a multiple cloning site between the proteins, the TM3–4 loops of different subunits or the basic motif 346RRKRR350 were inserted in frame. B, shown is confocal live fluorescence microscopy of HEK293 cells transfected with fusion proteins of GFP and β-galactosidase (β-gal-GFP, 153 kDa), the TM3–4 loop of hsGlyRα1 with GFP and β-galactosidase ((β-gal)-α1-GFP, 162 kDa), and the basic motif 346RRKRR350 with GFP and β-galactosidase ((β-gal)-RRKRR-GFP, 153 kDa). Although (β-gal)-GFP is excluded from the nucleus, (β-gal)-α1-GFP is actively imported into the nucleus. The isolated basic motif RRKRR, however, is not sufficient to mediate nuclear import. C, shown is confocal live fluorescence microscopy of HEK293 cells transfected with fusion proteins of GFP, β-galactosidase, and the TM3–4 loops of human GlyRα2 ((β-gal)-α2-GFP), GlyRα3K ((β-gal)-α3K-GFP), GlyRα3L ((β-gal)-α3L-GFP), and GlyRβ ((β-gal)-β-GFP). Fusion proteins (β-gal)-α1-GFP, (β-gal)-α3L-GFP, and (β-gal)-α3K-GFP, but not (β-gal)-α2-GFP and (β-gal)-β-GFP, are actively imported into the nucleus.
Detection of Full-length Nuclear GlyR
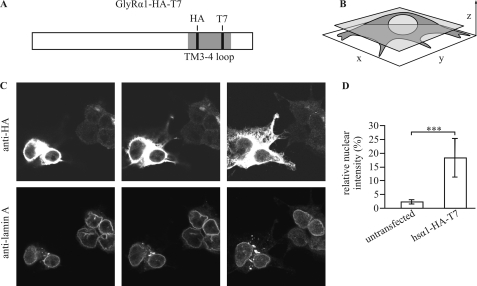
The detection of low amounts of nuclear GlyR antigen using monoclonal antibodies mAb4a or mAb2b directed against the N-terminal domain of all GlyRα subunits (mAb4a) or GlyRα1 (mAb2b), respectively, was hindered by considerable nonspecific nuclear reactivity of the antibodies (data not shown). To improve the signal-to-background ratio and to facilitate direct detection of the intracellular domain, we introduced small epitope tags into the TM3–4 loop. To conserve structure and function of the receptor, we inserted short and uncharged HA epitope and T7 major capsid protein epitope tags into the variable regions (Fig. 1C) of the TM3–4 loop of the full-length GlyRα1 subunit, obtaining the construct GlyRα1-HA-T7. Subsequently, GlyRα1-HA-T7 was tested for expression, sorting, and function, revealing no gross differences from the wild-type counterpart (supplemental Fig. 1).
Construct GlyRα1-HA-T7 was used to detect nuclear reactivity with confocal immunofluorescence-tomography (Fig. 6). To clearly visualize the boundaries of the nucleus, co-staining with an antibody directed against lamin A, a protein enriched in the nuclear envelope, was performed. In section-planes parallel to the coverslip through medial cellular levels, nuclear regions of interest medial to the anti-lamin A peak-immunoreactivity were marked, and the relative fluorescence intensity of anti-HA immunoreactivity was quantified. Nuclear immunoreactivity for the GlyR-HA-T7 antigen was significantly higher in transfected (18.3 ± 7.0%) as compared with untransfected HEK293 cells (2.2 ± 0.9%), indicating that nuclear import occurs upon transfection of the full-length receptor.
FIGURE 6.
Nuclear GlyR detection with a dual affinity tag. A, schematic representation of the construct used (hsGlyRα1-HA-T7) is shown; note the full-length hsGlyRα1 (white box) carrying both a T7 and an HA tag (black lines) within the TM3–4 loop (gray bar). B, visualization is shown of the orientation of tomography with a representative medial x-y section plane (transparent gray). C, shown is confocal immunofluorescence of HEK293 cells transfected with hsGlyRα1-HA-T7 and stained using anti-HA- (upper panels) and anti-lamin A antibody (lower panels). HA reactivity was measured at high laser intensities to enable detection of few nuclear epitopes. Two representative transfected cells (left) and four representative barely or un-transfected cells (right) within one visual field are depicted. Selection of section planes parallel to the coverslip (x-y) ranging from the adhesive part (right panel) to the highest level of the cells (left panel) is shown. D, shown is quantification of nuclear intensities using anti-HA-antibody from tomographies of 172 untransfected and 62 hsα1-HA-T7-transfected cells. Significance is calculated with t test resulting in p < 0.001 (***). A fraction of the GlyR is imported into the nucleoplasm of transfected HEK293 cells.
Even though full-length receptor constructs were transfected, it is possible that the nuclear immunoreactivity observed in confocal immunofluorescence-tomography stems from fragment peptides that are generated by posttranslational cleavage of the subunit. To show that the observed nuclear immunoreactivity corresponds to the full-length receptor, it has to be ruled out that fragments are generated that hold the NLS and, thus, might also be imported into the nucleus.
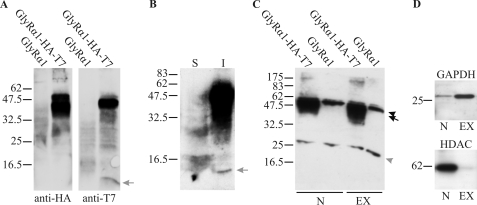
In Western blot analysis of lysate from GlyRα1-HA-T7-transfected HEK293 cells, we detected a fragment of about 9 kDa (Fig. 7A). This fragment was stained only with the antibody against the T7 tag but not with the antibody against the HA tag, indicating probable cleavage between the tags in the central part of the TM3–4 loop. Such a cleavage would generate a fragment of 7.5–12 kDa that holds TM4 but not the NLS as the NLS is N-terminal of the HA tag (supplemental Figs. 1A). To further corroborate the identity of this fragment, we prepared a membrane fraction from transfected HEK293 cells using alkaline extraction, pH 11.5. This method is used to distinguish between membrane-associated and membrane-integral proteins. A protein that cannot be solubilized from the membrane fraction by alkaline extraction is generally regarded to be a membrane integral protein, e.g. with a transmembrane domain. Consistent with the size of the fragment and the site of the cleavage, the fragment was not soluble after extraction, suggesting that, indeed, it harbors TM4 (Fig. 7B). Altogether, these results show that the nuclear immunoreactivity observed in confocal microscopy probably did not correspond to this fragment.
FIGURE 7.
Probing the proteolytic integrity of nuclear GlyRs. A, shown is a Western blot with anti-HA (left) and anti-T7 antibodies (right). Whole-cell lysates are shown of HEK293 cells transfected with human GlyRα1 (lanes 1 and 3) and GlyRα1-HA-T7 (lanes 2 und 4, see the scheme in Fig. 6A). The expected molecular mass for the full-length receptor is ∼49 kDa. A low molecular mass fragment generated from GlyRα1-HA-T7 is marked (gray arrow). As this fragment is only recognized by the anti-T7 antibody and not the anti HA antibody, it corresponds to a cleavage between these epitopes (compare Fig. 6A). B, alkaline extraction of membrane preparations of hsGlyRα1-HA-T7-transfected HEK293 cells is shown. Soluble (S) and insoluble (I) proteins after extraction are shown. The low molecular mass fragment (gray arrow) is insoluble after alkaline extraction and, thus, as expected, holds TM4. C, immunoprecipitation with anti-T7-antibody from nuclear (N) or extranuclear (EX) compartments of HEK293 cells transfected with hsGlyRα1 (lanes 2 and 4) or hsGlyRα1-HA-T7 (lanes 1 and 3). In this Western blot with anti-HA-antibody the heavy (upper arrowhead) and the light chain (lower arrowhead) of the antibody and the unprocessed hsGlyRα1-HA-T7 (black arrow) are marked. D, controls for enrichment of different compartments with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (extranuclear) and anti-histone deacetylase 1 (HDAC) antibody (nuclear). Taken together, these results exclude that significant amounts of soluble fragments of the GlyR are generated in transfected HEK293 cells.
To conclude that the observed nuclear immunoreactivity resulted from full-length subunits, it is essential to rule out that such a soluble fragment was generated as a potential soluble fragment might be easily imported into the nucleus. Although no such fragment was detected in high sensitivity Western blot analysis, we attempted to even further enhance analytical sensitivity. An assay was designed to detect even very low amounts of soluble TM3–4 loop fragments that would be imported into the nucleus. This was achieved by nuclear compartmental extraction, subsequent immunoprecipitation with an antibody directed against the T7-tag, and Western blot analysis using an antibody directed against the HA tag (Fig. 7, C and D). Importantly, the full-length subunit, but no immunolabeled fragment, was detectable even with extended exposure times, indicating that the nuclear epitopes in immunocytochemistry solely represent full-length GlyRs. It should be noted that most of the precipitated full-length GlyRs in the nuclear compartment are probably ER contaminants. Because the outer nuclear membrane is continuous with the rough endoplasmic reticulum, it cannot be separated fully from nuclear compartmental extractions. This leads to an overestimation of the amount of nuclear GlyR if judged by band intensity in the Western blot. Nevertheless, together with immunocytochemical data, which clearly distinguish between nuclear envelope and nucleoplasmic staining (Fig. 6), it can be concluded that the minor intranuclear staining observed with the HA antibody represents full-length subunits.
Karyopherins Interact with the TM3–4 Loop
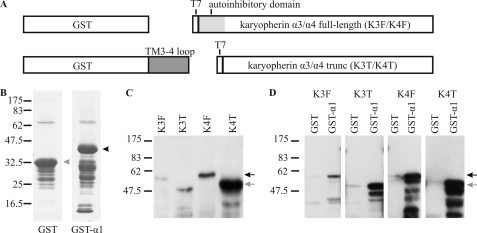
In an independent yeast two-hybrid assay, we screened for potential interaction partners of the TM3–4 loop of rnGlyRα3L. Of 71 positive clones, 51 corresponded to either karyopherin α3 or α4 (data not shown). Karyopherin α-subunits bind to karyopherin β1 via their autoinhibitory domains, thereby opening their NLS binding pockets. This pocket holds multiple, partially overlapping binding sites for NLSs. Trimeric complexes of α-karyopherins, karyopherin β1, and NLS-harboring proteins are transferred through the nuclear pore complex in an energy-dependent process. In vitro, in the absence of free karyopherin β1, the affinity of α-karyopherins to their cargo is reduced by 3 orders of magnitude (25). For GST pulldown assays, we therefore generated full-length and truncated karyopherin α3 and α4 lacking the N-terminal autoinhibitory domain (Fig. 8A). All four constructs were pulled down with GST fused to the TM3–4 loop of GlyRα1 but not with GST alone (Fig. 8, B–D). This indicates a high in vitro affinity of GlyRα1 to karyopherin α3 and α4. Taken together, the yeast two-hybrid assay and GST pulldown detected molecular interactions for the TM3–4 loops of rnGlyRα1, -α3L, and -α3K but not for α2 and β. For GlyRα3L, these interactions were found to be dependent on the basic NLS motif.
FIGURE 8.
GST pulldown assay of karyopherins α3 and α4. A, bars represent the constructs used. B, shown is a Coomassie gel after purification of GST (gray arrowhead) and GST fused to the TM3–4 loop of human GlyRα1 (GST-α1, black arrowhead). C, shown is a Western blot with anti-T7-antibody after bacterial expression of karyopherin α3 (K3) und α4 (K4) with a T7 tag. Full-length (F, black arrow) and N-terminal-truncated (T, gray arrow) constructs lacking the autoinhibitory domain are shown. D, Western blot with an anti-T7-antibody detecting full-length and truncated karyopherins pulled down with GST and GST-α1. Both karyopherins specifically bind to the TM3–4 loop in vitro. Binding is not hindered by the presence of the autoinhibitory domain.
To test the possibility of GlyR-karyopherin interactions in neurons, we performed confocal immunocytochemistry of spinal cord neuronal cultures using antibodies against GlyRα1 (mAb2b), synaptophysin (as synaptic marker) and karyopherin α3 or α4 (data not shown). Only minimal association of GlyRα1 and karyopherins α3 or α4 was observed, indicating that there is no strong static interaction in vivo. Nevertheless, this observation does not rule out the presence of a potentially short-lived complex between karyopherins and the GlyR.
DISCUSSION
Structural comparisons of GlyR subunit polypeptides showed that a conserved basic motif is located within the TM3–4 loop of GlyRα1 and -α3, whereas the homologous sequence varies slightly in the α2 and β subunits. Sequence analysis of the basic motif present in the GlyRα1 and -α3 subunits predicted this sequence to correspond to a nuclear localization signal. However, basic motifs may be functionally ambiguous; in addition to the nuclear sorting of proteins, the basic motif present in the TM3–4 loops of GlyRα1 and -α3 may be multifunctional and also mediate the interaction with a molecular chaperone (26). When tested for functionality in HEK293 cells and primary hippocampal neurons, the intracellular domain of the GlyR was, indeed, subject to nuclear sorting.
To achieve nuclear sorting, the basic motif 346RRKRR350 in the conserved intracellular region C1 was required but not sufficient. As shown by the use of large molecular weight constructs that restrict diffusion through the nuclear pore, nuclear import was mediated by functional NLS motifs in GlyRα1 and -α3 subunits. A similar trafficking process has been observed for the N-methyl-d-aspartate receptor variant NR1. The intracellular domain of NR1 is imported into the nucleus via a functional NLS, as analyzed by recombinant expression of GFP fusion constructs (27). Likewise, the isolated intracellular domain constructs of the receptor-tyrosine kinase notch (28) and the L-type voltage-gated calcium channel CaV1.2 (29) are subject to nuclear sorting, indicating that various types of transmembrane proteins harbor nuclear trafficking motifs. For these proteins, however, the proportion of nuclear antigen generated from recombinant or endogenous full-length proteins was low, demonstrating that the detection of nuclear antigen originating from transmembrane proteins requires sensitive detection techniques (27, 29, 30).
Indeed, the detection of nuclear GlyR antigen by monoclonal antibodies (mAb2b, mAb4a) was limited by low signal to background ratios for nuclear staining. To enhance sensitivity and to allow for detection of intracellular domain fragments, we introduced affinity tags into the TM3–4 loop of the full-length GlyRα1 subunit. Using high laser intensities in confocal imaging, we succeeded in the detection of nucleoplasmic immunoreactivity when HEK293 cells were transfected with full-length tagged GlyR constructs, indicating nuclear import.
The transmembrane proteins NR1, notch, and CaV1.2 undergo proteolytic cleavage before their intracellular domains are translocated into the nucleus (27, 29, 31). Making use of affinity tags, we analyzed the proteolytic integrity of transfected GlyR constructs in HEK293 cells. A fragment was identified carrying the T7 tag in the C-terminal part of the TM3–4 loop, indicating proteolytic cleavage. As deduced from the apparent molecular weight and resistance to alkaline extraction, this fragment most likely also held the TM4 region as well as the C terminus of the subunit. The proteolytic cleavage probably occurred between the conserved regions C1 and C3 of the TM3–4 loop. This localization is consistent with a study reporting ubiquitination and subsequent cleavage of a C-terminal-tagged GlyR construct expressed in Xenopus oocytes (32). The T7-tagged fragment we observed, however, neither harbored the basic motif 346RRKRR350 essential for nuclear import nor did it hold the further N-terminal HA tag used for detection of the GlyR in the nucleus. Thus, it could not have contributed to the observed nucleoplasmic GlyR immunoreactivity. As detection of nuclear soluble fragments might have been limited by sensitivity, we enriched the antigen by nuclear compartmental extraction and immunoprecipitation. Still, we found no evidence for a fragment containing both affinity tags, whereas full-length GlyR subunits were readily detectable in the nuclear compartment of transfected HEK293 cells. These results indicate that the epitopes present in the nucleus corresponded to full-length GlyR subunits.
Nuclear import relies on an intricate cellular machinery, where translocation of NLS harboring proteins is frequently mediated by karyopherins. Indeed, interaction of the GlyR intracellular domains with karyopherins α3 and α4 was strong in in vitro studies involving yeast two-hybrid screening and GST pulldown assays. Immunocytochemistry of primary neuronal cultures, however, did not indicate a strong interaction in vivo. Nevertheless, the interaction of karyopherins with the TM3–4 loops of GlyRα1 and -α3, but not of GlyRα2 and -β, closely resembled the subunit specificity of the nuclear import in our reporter systems. This suggests that the interaction with karyopherins underlies the observed nuclear import of the TM3–4-loop constructs. Besides, the specificity of karyopherin interaction may represent one of the mechanisms resulting in differential trafficking of the GlyR subunits. The basic motif 346RRKRR350 was required but not sufficient to mediate nuclear sorting. Resembling the situation for NR1 (27), the interaction motif located within the TM3–4 loop might, therefore, be a bipartite NLS, potentially also involving basic residues in C3.
In addition to nuclear import, extranuclear functions of the interaction between GlyRs and karyopherins are conceivable. Indeed, α-karyopherins have been described to mediate transport functions in pre- and postsynaptic compartments of neurons (33–36). It has been proposed that karyopherin-mediated signaling from the synapse to the nucleus represents a general mechanism for regulation of synaptic plasticity (37). The GlyR might, therefore, serve as a synaptic anchor liberating karyopherins upon channel opening and chloride influx. Eventually, karyopherin α3 and α4 might mediate the interaction of GlyRs to the dynein transport complex via karyopherin β1, representing a second link in addition to gephyrin (38, 39).
The basic motif RRKRR has been shown to be part of an important determinant of GlyRα1 topology, namely RXRRKRR, in oocyte systems (40). The authors concluded that the basic residues are required to counteract the propensity for intracellular retention of the TM2–3 loop resulting from three basic amino acids therein. Recently, we showed that RRKRR, as a part of RRKRRH, greatly enhances the potential of truncated oscillator GlyRα1 subunits to be rescued with tail domains, indicating an important role in protein-protein interaction (41). Here, we present the interaction with karyopherins and the resulting regulation of intracellular sorting as a novel role for this multifunctional basic motif. Consistent with its multifunctional role, the basic motif may also mediate interaction to other proteins such as the molecular chaperone VCP/p97, which has been reported to bind short, basic motifs (26). These interactions might represent the molecular mechanism underlying both correct membrane topology and efficient functional rescue. Furthermore, sequence diversity within this motif, as seen for GlyRα2, may underlie differential regulation of single activities of GlyR subunit variants.
Supplementary Material
Acknowledgments
We thank Drs. Martin Eberhardt, Stephan Hupfer, and Katrin Schiebel for critical reading of this manuscript and for helpful comments. Technical assistance of Yvonne Pechmann, Rosa Weber, and Marina Wenzel is gratefully acknowledged. We thank Dr. Julia White (GlaxoSmithKline) for assistance with yeast two-hybrid screening.
This work was supported by Deutsche Forschungsgemeinschaft (BE1138/5-3), Elitenetzwerk Bayern (Lead Structures of Cell Function, K-BM-2003-85), the Medical Research Council (G0500833, G0501258, and G0601585 to K. H. and R. J. H.), and European Framework Program 7 (Neuro Cypress, HEALTH-F4-2008-202088).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- GlyR
- glycine receptor
- TM
- transmembrane domain
- NLS
- nuclear localization signal
- hs
- H. sapiens
- mm
- M. musculus
- rn
- R. norvegicus
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 2.Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 3.Breitinger H. G., Becker C. M. (2002) Chembiochem. 3, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 4.Flicek P., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Eyre T., Fitzgerald S., Fernandez-Banet J., Gräf S., Haider S., Hammond M., Holland R., Howe K. L., Howe K., Johnson N., Jenkinson A., Kähäri A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A. J., Vogel J., White S., Wood M., Birney E., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Kasprzyk A., Proctor G., Smith J., Ureta-Vidal A., Searle S. (2008) Nucleic Acids Res. 36, D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker C. M., Hoch W., Betz H. (1988) EMBO J. 7, 3717–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kneussel M., Betz H. (2000) Trends Neurosci. 23, 429–435 [DOI] [PubMed] [Google Scholar]
- 7.Grudzinska J., Schemm R., Haeger S., Nicke A., Schmalzing G., Betz H., Laube B. (2005) Neuron 45, 727–739 [DOI] [PubMed] [Google Scholar]
- 8.Kim E. Y., Schrader N., Smolinsky B., Bedet C., Vannier C., Schwarz G., Schindelin H. (2006) EMBO J. 25, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lévi S., Logan S. M., Tovar K. R., Craig A. M. (2004) J. Neurosci. 24, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saiyed T., Paarmann I., Schmitt B., Haeger S., Sola M., Schmalzing G., Weissenhorn W., Betz H. (2007) J. Biol. Chem. 282, 5625–5632 [DOI] [PubMed] [Google Scholar]
- 11.Fritschy J. M., Harvey R. J., Schwarz G. (2008) Trends Neurosci. 31, 257–264 [DOI] [PubMed] [Google Scholar]
- 12.Maas C., Tagnaouti N., Loebrich S., Behrend B., Lappe-Siefke C., Kneussel M. (2006) J. Cell Biol. 172, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bluem R., Schmidt E., Corvey C., Karas M., Schlicksupp A., Kirsch J., Kuhse J. (2007) J. Biol. Chem. 282, 37783–37793 [DOI] [PubMed] [Google Scholar]
- 14.Yevenes G. E., Moraga-Cid G., Guzmán L., Haeger S., Oliveira L., Olate J., Schmalzing G., Aguayo L. G. (2006) J. Biol. Chem. 281, 39300–39307 [DOI] [PubMed] [Google Scholar]
- 15.The UniProt Consortium: The Universal Protein Resource (UniProt) (2008) Nucleic Acids Res. 36, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 17.Croci C., Brändstatter J. H., Enz R. (2003) J. Biol. Chem. 278, 6128–6135 [DOI] [PubMed] [Google Scholar]
- 18.Oertel J., Villmann C., Kettenmann H., Kirchhoff F., Becker C. M. (2007) J. Biol. Chem. 282, 2798–2807 [DOI] [PubMed] [Google Scholar]
- 19.Garg A., Bhasin M., Raghava G. P. (2005) J. Biol. Chem. 280, 14427–14432 [DOI] [PubMed] [Google Scholar]
- 20.Horton P., Nakai K. (1997) Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 147–152 [PubMed] [Google Scholar]
- 21.Cokol M., Nair R., Rost B. (2000) EMBO Rep. 1, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brameier M., Krings A., MacCallum R. M. (2007) Bioinformatics 23, 1159–1160 [DOI] [PubMed] [Google Scholar]
- 23.Sorg G., Stamminger T. (1999) Biotechniques 26, 858–862 [DOI] [PubMed] [Google Scholar]
- 24.Nikolic Z., Laube B., Weber R. G., Lichter P., Kioschis P., Poustka A., Mülhardt C., Becker C. M. (1998) J. Biol. Chem. 273, 19708–19714 [DOI] [PubMed] [Google Scholar]
- 25.Fanara P., Hodel M. R., Corbett A. H., Hodel A. E. (2000) J. Biol. Chem. 275, 21218–21223 [DOI] [PubMed] [Google Scholar]
- 26.Boeddrich A., Gaumer S., Haacke A., Tzvetkov N., Albrecht M., Evert B. O., Müller E. C., Lurz R., Breuer P., Schugardt N., Plassmann S., Xu K., Warrick J. M., Suopanki J., Wüllner U., Frank R., Hartl U. F., Bonini N. M., Wanker E. E. (2006) EMBO J. 25, 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes K. D., Mattar P., Marsh D. R., Jordan V., Weaver L. C., Dekaban G. A. (2002) J. Neurochem. 81, 1152–1165 [DOI] [PubMed] [Google Scholar]
- 28.Lieber T., Kidd S., Alcamo E., Corbin V., Young M. W. (1993) Genes Dev. 7, 1949–1965 [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Ospina N., Tsuruta F., Barreto-Chang O., Hu L., Dolmetsch R. (2006) Cell 127, 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struhl G., Adachi A. (1998) Cell 93, 649–660 [DOI] [PubMed] [Google Scholar]
- 31.Fiúza U. M., Arias A. M. (2007) J. Endocrinol. 194, 459–474 [DOI] [PubMed] [Google Scholar]
- 32.Büttner C., Sadtler S., Leyendecker A., Laube B., Griffon N., Betz H., Schmalzing G. (2001) J. Biol. Chem. 276, 42978–42985 [DOI] [PubMed] [Google Scholar]
- 33.Dieterich D. C., Karpova A., Mikhaylova M., Zdobnova I., König I., Landwehr M., Kreutz M., Smalla K. H., Richter K., Landgraf P., Reissner C., Boeckers T. M., Zuschratter W., Spilker C., Seidenbecher C. I., Garner C. C., Gundelfinger E. D., Kreutz M. R. (2008) PLoS Biol. 6, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson K. R., Otis K. O., Chen D. Y., Zhao Y., O'Dell T. J., Martin K. C. (2004) Neuron 44, 997–1009 [DOI] [PubMed] [Google Scholar]
- 35.Yudin D., Hanz S., Yoo S., Iavnilovitch E., Willis D., Gradus T., Vuppalanchi D., Segal-Ruder Y., Ben-Yaakov K., Hieda M., Yoneda Y., Twiss J. L., Fainzilber M. (2008) Neuron 59, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanz S., Perlson E., Willis D., Zheng J. Q., Massarwa R., Huerta J. J., Koltzenburg M., Kohler M., van-Minnen J., Twiss J. L., Fainzilber M. (2003) Neuron 40, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 37.Otis K. O., Thompson K. R., Martin K. C. (2006) Curr. Opin. Neurobiol. 16, 329–335 [DOI] [PubMed] [Google Scholar]
- 38.Lischka P., Sorg G., Kann M., Winkler M., Stamminger T. (2003) J. Virol. 77, 3734–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlson E., Hanz S., Ben-Yaakov K., Segal-Ruder Y., Seger R., Fainzilber M. (2005) Neuron 45, 715–726 [DOI] [PubMed] [Google Scholar]
- 40.Sadtler S., Laube B., Lashub A., Nicke A., Betz H., Schmalzing G. (2003) J. Biol. Chem. 278, 16782–16790 [DOI] [PubMed] [Google Scholar]
- 41.Villmann C., Oertel J., Ma-Högemeier Z. L., Hollmann M., Sprengel R., Becker K., Breitinger H. G., Becker C. M. (2009) J. Neurosci. 29, 2440–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.