Abstract
The stigmatic estolide is a lipid-based polyester constituting the major component of exudate in solanaceous plants. Although the exudate is believed to play important roles in the pollination process, the biosynthetic pathway of stigmatic estolide, including genes encoding the key enzymes, remains unknown. Here we report the cloning and characterization of the cytochrome P450 gene CYP86A22, which encodes a fatty acyl-CoA ω-hydroxylase involved in estolide biosynthesis in the stigma of Petunia hybrida. A CYP86A22 cDNA was isolated from a developing stigma cDNA library, and the corresponding gene was shown to express predominantly in the developing stigma. Among six P450 genes isolated from this library, only CYP86A22 was implicated in ω-hydroxylation following RNA interference (RNAi)-mediated suppression. Unlike wild-type plants in which ω-hydroxy fatty acids (mainly in the form of 18-hydroxy oleic acid and 18-hydroxy linoleic acid) compose 96% of total stigma fatty acids, the ω-hydroxy fatty acids were essentially absent in the stigmas from 18 of 46 CYP86A22-RNAi transgenic plants and had varying levels of suppression in the remaining 28 plants. Furthermore, lipids in the 18 CYP86A22-RNAi stigmas were predominantly triacylglycerols and diacylglycerols instead of the estolides, which characterize the wild-type stigma. Analyses of recombinant CYP86A22 conclusively demonstrated that this P450 is a ω-hydroxylase with a substrate preference for both saturated and unsaturated acyl-CoAs rather than free fatty acids. We conclude that the cytochrome P450 enzyme CYP86A22 is the key fatty acyl-CoA ω-hydroxylase essential for the production of ω-hydroxy fatty acids and the biosynthesis of triacylglycerol-/diacylglycerol-based estolide polyesters in the petunia stigma.
Keywords: Cytochromes/Cytochrome P450, Lipid/Fatty Acid, Lipid/Triacylglycerol, Hydroxylase, siRNA, Acyl-Coenzyme A, Estolide Polyester, Fatty Acid Hydroxylation, Petunia Flower
Introduction
The stigma of Petunia hybrida and other solanaceous species is covered by copious exudate, whereas the stigma of Arabidopsis thaliana and other Brassicaceae has little or no exudate on the surface (1, 2). The exudate is primarily lipidic, although other compounds such as proteins, sugars, and pigments are also present (3–6). The lipidic character of the exudate derives from the presence of a large amount of estolide, a lipid-based polyester. Estolide is rich in ω-hydroxy fatty acids, i.e. fatty acids containing a hydroxy group on the terminal or omega (ω) carbon. The exudate of petunia stigma contains up to 96% ω-hydroxy fatty acids mainly in the form of 18-hydroxy-oleic acid and 18-hydroxy-linoleic acid (7, 8). These ω-hydroxy fatty acid monomers are thought to be attached to a glycerol backbone and then iteratively esterified such that the ω-hydroxyl group of one fatty acid is esterified to a carboxyl group of another to ultimately form the estolide. In tobacco stigma, estolides are formed as tetra-, penta-, hexa-, and heptaacyl glycerides and capped by normal fatty acids (9, 10). The stigmatic exudate has shown significant physiological roles in the pollination process (11–14). Despite its importance, the biosynthetic pathway of stigmatic estolide, including genes encoding the key enzymes, has not been elucidated.
Analyses of Arabidopsis cuticle mutants indicate that cytochrome P450 monooxygenases (CYPs or P450s)5 play a central role in the synthesis of cutin and suberin, two other major types of plant lipid-derived polyesters (15, 16). P450s are hemoproteins belonging to a large superfamily virtually ubiquitous in all living organisms (17, 18). The majority of microsomal P450s studied to date require a cognate NADPH-dependent reductase to transfer electrons from NADPH via FAD and FMN cofactors to the prosthetic heme group of the P450s. Interestingly, unlike yeast and animal systems, which typically rely on a single P450 reductase, plants appear to contain two distinct P450 reductases (19, 20). P450s catalyze a wide range of biological oxidations, including fatty acid ω-hydroxylations. Among various organisms, these fatty acid ω-hydroxylases are grouped into divergent P450 families. The mammalian P450s mediating fatty acid ω-hydroxylation are predominately found in the CYP4 family (21–23), whereas the corresponding fungal P450s are in the CYP52 family (24, 25). In plants, these P450s are usually found in the CYP86 (26, 27) and CYP94 families (28, 29).
Genes encoding P450 fatty acid ω-hydroxylases have been isolated from A. thaliana and other plant species. The plant P450 fatty acid ω-hydroxylases characterized to date have been implicated almost exclusively in cutin and suberin synthesis. For example, the first isolated cutin mutant, att1 encoding cyp86a2 from A. thaliana, exhibits a reduction of cutin by 70% (30). The Arabidopsis cyp86a8 mutant displays severely postgenital organ fusion resulting from a structural defect at the cuticle layer (31). In vitro assays indicate that the recombinant CYP86 enzymes such as CYP86A1, CYP86A2, CYP86A4, CYP86A7, and CYP86A8 all have the ability to ω-hydroxylate free fatty acids with chain lengths from C12 to C18 (26, 27), excluding stearic acid (18:0) and 9,10-epoxystearic acid (31, 32). CYP94A subfamily members also include fatty acid ω-hydroxylases; however, these P450s have a substrate preference distinct from the CYP86A members. For instance, CYP94A1 from Vicia sativa is able to ω-hydroxylate not only free fatty acids with chain lengths from C10 to C18 (not including 18:0) but also 9,10-dihydroxystearic and 9,10-epoxystearic acid (28). CYP94A5 from Nicotiana tobacum utilizes fatty acids with chain lengths similar to the above P450s but preferentially uses 9,10-epoxystearic acid to produce 18-hydroxy-9,10-epoxystearic acid, one of the major C18 cutin monomers (29). Collectively, the P450 fatty acid ω-hydroxylases implicated in the cutin biosynthetic pathway all appear to use free fatty acids as substrates, and to date, no P450 hydroxylases that utilize fatty acyl-CoAs for the synthesis of extracellular lipids have been reported in plants.
In this study, we report the cloning and characterization of the cytochrome P450, CYP86A22, a fatty acyl-CoA ω-hydroxylase isolated from the stigma of P. hybrida. Our data demonstrate that CYP86A22 is the P450 enzyme responsible for the synthesis of ω-hydroxy fatty acids in the petunia stigma. Furthermore, our results establish that CYP86A22 activity is an essential prerequisite for the biosynthesis of the DAG- and TAG-based estolide polyesters that constitute the bulk of the stigmatic exudate. Our data suggest that the CYP86A22 has substrate specificity distinct from the other P450 fatty acid ω-hydroxylases reported to date in which recombinant CYP86A22 exhibits a clear preference for both saturated and unsaturated fatty acyl-CoAs rather than free fatty acids. Taken together, the cytochrome P450 enzyme CYP86A22 is identified as the key fatty acyl-CoA ω-hydroxylase essential for estolide synthesis in the petunia stigma.
EXPERIMENTAL PROCEDURES
Materials
[1-14C]Palmitic acid (55 mCi/mmol), [1-14C]oleic acid (53 mCi/mmol), and [1-14C]stearic acid (53 mCi/mmol) were purchased from American Radiolabeled Chemicals (St. Louis, MO). The corresponding [1-14C]acyl-CoAs were either purchased from American Radiolabeled Chemicals or synthesized as described by King et al. (33). All other chemicals were obtained from Sigma unless indicated otherwise.
Seeds of P. hybrida were purchased from Burpee (Warminster, PA). Plants were grown in a greenhouse at 22 °C under 15 h of daylight at 50–60% relative humidity. In this study, floral development was divided into four stages: stage 1, floral buds with a length of 1 cm; stage 2, floral buds with a length of 2 cm; stage 3, floral buds with a length of 3 cm; and stage 4, floral buds with a length of 4 cm.
Cloning of Cytochrome P450 Hydroxylase and NADPH-cytochrome P450 Reductase Genes
One milligram of total RNA was extracted from stage 2 developing stigmas of petunia floral buds using the RNeasy Plant Kit (Qiagen). The poly(A)+-enriched mRNA was purified from total RNA, and a cDNA library, which contained 2.9 × 107 primary plaques, was constructed in the Uni-Zap II vector by Stratagene (La Jolla, CA). To identify clones encoding cytochrome P450 monooxygenases and NADPH-cytochrome P450 reductases, an expressed sequence tag (EST)-based approach was used. Plasmid DNA (pBluescript SK−) was prepared via in vivo mass excision according to Stratagene's protocols. ESTs were produced from a total of 8,737 high quality 5′ sequences of the plasmids and then annotated using results from a tBlastn search of GenBankTM. Forty-one ESTs were identified to unambiguously align to established cytochrome P450s. These ESTs were further classified into six distinct groups. To obtain full-length cDNA clones, digoxigenin (DIG)-labeled (Roche Applied Science) probes were generated by PCR using a cDNA clone from the corresponding EST group as a template. The oligonucleotide primers used for PCR were: the forward primer, 5′-CGACACGTGGCTTTTCCAAC-3′, and the reverse primer, 5′-AACTCTGGAGAGAGTGTCTC-3′, for PH1 (CYP86A22) probe; the forward primer, 5′-ATGATGATAGCAATTATCTTAG-3′, and the reverse primer, 5′-TGCTAAGAATTTCCTTGATC-3′, for PH3 (CYP749B1) probe; the forward primer, 5′-TGGAAGATTAAGAAAGCTC-3′, and the reverse primer, 5′-CTATCTGTGAAAGATACG-3′, for PH5 (CYP704A14) probe; and the forward primer, 5′-TGGATTAGTATCTTTCTTC-3′, and the reverse primer, 5′-AGCAAATCCAGGGATATTG-3′ for PH6 (CYP88C1) probe. Each DIG-labeled probe was used to screen the petunia stigma cDNA library following the recommended protocol (Roche Applied Science). For both PH2 (CYP714G3) and PH4 (CYP94A13), sequence analyses indicated that the primary EST clones encoded full-length cDNAs. All full-length cDNA clones were verified by sequencing both strands.
Our sequence analysis also revealed two EST-encoding proteins with homology to established NADPH-P450 reductases (PRs). To isolate a clone encoding the full-length cDNA for the putative reductase PR1, we again screened our stigma cDNA library using a DIG-labeled probe. The probe was generated by PCR using the PR1 EST-specific primers, 5′-ACCATCCTTGCAAAGTAAATG-3′ and 5′-TGCTTCACTAGGTTCAGCTG-3′. As with the putative P450 (CYP714G3 and CYP94A13) ESTs, the EST clone for PR2 appeared to encode a full-length cDNA. The sequences of PR1 and PR2 cDNAs were verified as above.
To clone the CYP86A22 promoter, an inverse PCR was performed using petunia genomic DNA as template. One microgram of genomic DNA was digested with 10 units of either NcoI or SpeI restriction enzymes. The resulting DNA fragments were diluted to a final concentration of 3–4 ng/μl and self-ligated by T4 DNA ligase (New England Biolabs, Beverly, MA). The circularized products were purified and used as template for PCR amplification. Primers specific to the 5′-end sequence of CYP86A22 cDNA were: forward primers, 5′-CGACACGTGGCTTTTCCAAC-3′ or 5′-GATGACTTGCTTTCGAGGTTC-3′, and a reverse primer, 5′-GCTGCTACAATTGCTACAATC-3′. The PCR was performed in a 50-μl reaction volume containing 20 pmol of each primer, 2.5 units of pfu DNA polymerase (Stratagene), 0.2 mm dNTP, 2 mm Mg2+, and 100 ng of the ligated DNA. The PCR conditions used were: one cycle of 4 min at 94 °C, 30 cycles of 30 s at 93 °C, 30 s at 55 °C, and 4 min at 72 °C. The amplified product was visualized on a 1% agarose gel, purified, and cloned into pCR-Blunt II-TOPO vector (Invitrogen), resulting in pCR-StigP.
Constructs for Plant and Insect Cell Expression
To generate a stigma expression vector, the CYP86A22 promoter sequence was used to replace the phaseolin promoter in Gateway binary vector pGATE-Phas (Dow AgroSciences, Indianapolis, IN), which contains the aadA and bar genes for selection in E. coli and plants, respectively. Because no restriction enzyme site is located at the 5′-end of phaseolin promoter, an SphI site was introduced by PCR. Four primers served this purpose: I, 5′- ATTCGGCGCCTACATCGACG-3′; II, 5′-AATTGCAGCCCCGGCCGCCAGCATGCTTGTACTCCCAGTATCATTATAG-3′; III, 5′-ATAATGATACTGGGAGTACAAGCATGCTGGCGGCCGGGGCTGCAATTG-3′; and IV, 5′-ACCTTAATTAAGGATCCTAGAGTAG-3′, in which the underlined sites are KasI, SphI, SphI, and PacI, respectively. Three continuous PCRs were performed. The first PCR was performed using primers I and III and pGATE-Phas as template, the second with primers II and IV and also pGATE-Phas as template, and the last using primers I and IV with the purified products from the first and second PCR reactions as templates. The final PCR product containing the added SphI site was purified and used to replace the KasI/PacI fragment in pGATE-Phas. The restriction sites SphI and PacI were introduced to the 5′- and 3′-ends of CYP86A22 promoter by PCR with primers 5′-AGCATGCAGAAAACTACAGTTTACTAATTTTTTTTCTAAG-3′ and 5′-AGTTAATTAACTTTTAATAACACCAACTTCCTG-3′, using the pCR-StigP as template. The CYP86A22 promoter was digested using SphI and PacI and cloned into the same sites in pGATE-Phas to generate pGATE-Stig vector, in which the phaseolin promoter is replaced by the CYP86A22 promoter that is upstream of a sense Gateway reading frame B (RfB).
We applied a Gateway (Invitrogen) cloning strategy to generate our multiple stigma-specific RNA interference (RNAi) constructs. Our stigma-specific Gateway RNAi destination vector is a pGATE-Stig derivative, in which the A. thaliana FAD2 intron1 is sandwiched between two RfB cassettes arranged as inverted repeats and cloned downstream of the CYP86A22 promoter. The FAD2 intron1 was amplified by PCR from A. thaliana genomic DNA using the forward primer, 5′-ACCCTGCAGGGTCCGTCGCTTCTCTTC-3′, and the reverse primer, 5′-ATGTTTAAACTGCAGAAAACCAAAAGC-3′. The SbfI and PmeI restriction enzyme sites are underlined. Similarly, the Gateway RfB was amplified by PCR from pGATE-Stig using primers 5′-ACCTCGAGATCAACAAGTTTGTACAAAAAAG-3′ and 5′-AGGTTTAAACATCAACCACTTTGTACAAG-3′, with the XhoI and PmeI underlined, respectively. The PCR fragments encoding the FAD2 intron (1.1 kb) and RfB (1.7 kb) were separately cloned into pCR-Blunt II-TOPO vector (Invitrogen). The 1.7-kb RfB fragment was then excised from the appropriate clone using PmeI and HindIII (vector) and cloned into the same sites of the appropriate vector containing the 1.1-kb FAD2 intron. Finally, the entire 2.8-kb fragment containing the FAD2 intron 1 and RfB was excised with SbfI and XhoI and inserted into the same sites of pGate-Stig, thereby placing the 2.8-kb fragment immediately downstream of the sense RfB cassette already present in the pGate-Stig vector. This also placed the FAD2 intron in the sense orientation and the second RfB cassette in antisense to the first, yielding a stigma-specific RNAi vector, pGate-Stig-RNAi, into which we could now easily introduce any interesting sequences using Gateway technology.
To evaluate the functions of putative P450s, RNAi-appropriate gene fragments were selected using RNAi searching tools (Dharmacon, Lafayette, CO). In an effort to reduce the possibility of spurious RNAi suppression, each sequence was aligned with multiple P450 cDNAs to identify those sequences containing regions of minimal overlap. The selected fragments (∼300–500 bp) were then amplified by PCR using the desired P450 cDNAs as a template. For TOPO directional cloning, four bases (CACC) were added to all forward primers where necessary. Gene-specific primers used for PCR were: 5′-CACCACCCGGACCTTGAG-3′ and 5′-TGTCACGTCCAGCTAGGATG-3′ for the CYP86A22 fragment, 5′-CACCTTCAAGAAGCTGCACTTATG-3′ and 5′-TATCACTTGTAAGATCGATG-3′ for the CYP714G3 fragment, 5′-CACCATGATGACAGCAATTTGC-3′ and 5′-GCTAAGAATTTCCTTGATC-3′ for the CYP749B1 fragment, 5′-CACCTAGTAAAATACCAACATCC-3′ and 5′-TTGCGCAAGGACCTAGAGT-3′ for the CYP94A13 fragment, 5′-CACCCTCATGTACAGGAGA-3′ and 5′-CTATCTGTGAAAGATACG-3′ for the CYP704A14 fragment, and 5′-CACCAAGGGCAAAGTCTAAC-3′ and 5′-CTCTGGGATAGTGTACC-3′ for the CYP88C1 fragment. Each PCR fragment was first cloned into the entry vector pENTR/D-TOPO (Invitrogen) and verified by sequencing. Each fragment was then inserted into the pGate-Stig-RNAi destination vector by recombination. The resulting vector contained an inverted repeat of the selected fragment spaced by the FAD2 intron1. Each pGate-Stig-RNAi construct was sequenced and then transferred into Agrobacterium tumefaciens for subsequent transformation into petunia plants. Petunia transformation was performed using the Agrobacterium-mediated leaf disc procedure (34). Transformants were regenerated from selective medium containing 15 mg/liter glufosinate ammonium (Finale, AgrEVo, Montvale, NJ). T1 lines were selected on soil containing 25 mg/liter glufosinate ammonium (Finale). T2 lines were grown in the same conditions.
The Bac-2-Bac system (Invitrogen) was used for heterologous expression in Spodoptera frugiperda (Sf-9) insect cells. To construct the necessary baculovirus expression vectors, CYP86A22, PR1, and PR2 were amplified by PCR using pfu DNA polymerase (Stratagene) and the appropriate cDNAs as a template. The forward and reverse primers were: 5′-AAAGGCCTATGGAAGTATCAACAACTATG-3′ and 5′-ATAGGCCTTCAAGCAGCAATCCCATTTAC-3′ for CYP86A22, 5′-ATCTCGAGATGGAGTCGAGTTCGTC-3′ and 5′-AAGCTAGCTCACCACACATCCCTGAG-3′ for PR1, 5′-ATCTCGAGATGGATTCTACAGCAGAAAAGC-3′ and 5′-TTGCTAGCTCACCACACATCACGCAG-3′ for PR2, respectively. The PCR products were cloned into the pCR-Blunt II-TOPO vector (Invitrogen) and verified by sequencing. The 1.7-kb CYP86A22 coding region was excised with StuI and cloned into the corresponding site of the pFastBac Dual expression vector (Invitrogen). Similarly, the 2.1-kb PR1 and PR2 coding regions were excised with XhoI and NheI and cloned into the corresponding sites in pFastBac (pFB) Dual. The resulting plasmids included single gene constructs, pFB-CYP86A22, pFB-PR1, and pFB-PR2, or double gene constructs, pFB-CYP86A22:PR1 and pFB-CYP86A22:PR2, which were then used to generate recombinant bacmids according to the recommended protocols. The empty pFB-Dual vector was used as a negative expression control. Typically, Sf-9 cells at a density of 1 × 106 cells/ml were infected with high titer virus and cultured for an additional 48 h. Cells were harvested by centrifugation and stored at −80 °C until the day of assay.
Histochemical GUS Analysis
The β-glucuronidase (GUS) coding sequence was amplified by PCR from pBI101.3 (Clontech, Mountain View, CA) using primers 5′-CACCTACGGTCAGTCCCTTATG-3′ and 5′-GGTAGCAATTCCCGAGGCTG-3′. The PCR product was cloned into the entry vector pENTR/D-TOPO (Invitrogen) and then inserted behind the stigma promoter in pGate-Stig vector. The resulting vector, pStig-GUS, was introduced into A. tumefaciens and used to transform petunia plants. Transgenic plants were selected by resistance to Basta and used for histochemical GUS analysis essentially as described by Jefferson et al. (35). Floral buds and tissues were removed and placed in the GUS substrate buffer (100 mm sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.5 mm X-Gluc). After a 5-min vacuum infiltration, samples were placed in a 37 °C incubator for 16 h. The tissues were then incubated in a solution of ethanol:acetic acid (3:1) at 70 °C for 1 h and rinsed twice with 70% ethanol. Blue signals were scored and photographed under a dissecting microscope.
Northern, Western, and 14C-Labeling Analysis
For Northern blot analysis, total RNA was isolated from the stigma and other tissues of petunia plants using the RNeasy Plant Kit (Qiagen). A total of 3 μg of RNA was separated on a 1.2% agarose gel containing 0.3 m formaldehyde and 0.25 μg of ethidium bromide. The RNA was subsequently transferred onto a nylon membrane and probed with the DIG-labeled gene-specific sequences. After hybridization, the signals were detected by a chemiluminescence reaction using an anti-DIG-alkaline phosphatase conjugate and the substrate disodium 3-(4-meth-oxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate.
For Western blot analysis, rabbit anti-CYP86A22 antisera was generated by Sigma-Genosys (Woodlands, TX) using the synthetic peptide SQDGDKFQVQDTFRFVA(C) corresponding to amino acids 435–451 of CYP86A22. Microsomal protein was extracted from the stigma of wild-type and transgenic plants as described by Benveniste et al. (36). Briefly, stigmas were ground in liquid nitrogen and extracted in a buffer containing 0.1 m potassium phosphate buffer (pH 7.4), 250 mm sucrose, 1 mm EDTA, 0.4% bovine serum albumin, and 10 mm 2-mercaptoethanol. The homogenate was centrifuged for 10 min at 10,000 × g to remove any tissue and cell debris. The resulting supernatant was then centrifuged at 500,000 × g for 15 min in an ultracentrifuge to pellet microsomal fraction. The microsomal fraction was washed once with the extraction buffer and then resuspended in a buffer consisting of 0.1 m potassium phosphate buffer (pH 7.4), 10 mm 2-mercaptoethanol, and 30% glycerol by gentle agitation with a glass rod. Protein concentrations were determined using the BCA assay (Pierce) with bovine serum albumin as a standard. Aliquots of the microsomes were then stored at −80 °C. Proteins were separated by SDS-PAGE on 4% stacking/10% resolving gel. Proteins were then electroblotted onto a nitrocellulose membrane and probed with a 1:1000 dilution of the CYP86A22 antisera followed by 1:4000 dilution of a goat anti-rabbit IgG alkaline phosphatase-conjugate (Sigma). Alkaline phosphatase was detected using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
For 14C-labeling analysis, two fresh stigmas were removed from the stage 2 floral buds of either wild-type or transgenic plants and placed in 0.1 ml of incubation buffer consisting of 25 mm sucrose, 100 mm sorbitol, and 25 mm MES, pH 6.0. One microcurie of [1-14C] 18:1 fatty acid (51 mCi/mmol) was then added to the buffer containing the two stigmas. After an overnight incubation under normal laboratory light, the samples were rinsed three times with water and briefly dried on a filter paper. Fatty acid methyl esters (FAMEs) from these stigmas were prepared as described below and separated by TLC on a silica gel 60 plate (EMD, Gibbstown, NJ) with a solvent system consisting of petroleum ether:diethyl ether:acetic acid (70:30:1, v/v). The radioactive signals were detected with an instant imager (Packard, Meriden, CT) and quantified using the manufacturer's software.
Enzyme Assays
To prepare microsomal fractions from Sf-9 cells, cell pellets were resuspended in 1× phosphate-buffered saline buffer containing 1 mm EDTA, 0.4 mm phenylmethylsulfonyl fluoride, and 10 mm 2-mercaptoethanol. Cells were then broken by manual homogenization in a 15-ml glass homogenizer on ice with 30 strokes. The lysate was centrifuged for 10 min at 10,000 × g to remove any cell debris. The resulting supernatant was then centrifuged at 500,000 × g for 15 min in an ultracentrifuge to pellet the microsomes. The microsomal fraction was washed twice with the lysis buffer to remove residual NADPH. The pellet was then resuspended in a buffer consisting of 1× phosphate-buffered saline buffer, 10 mm 2-mercaptoethanol, and 20% glycerol by gentle agitation with a glass rod. Protein concentrations were determined using the BCA assay (Pierce) with bovine serum albumin as a standard. Aliquots of the microsomes were stored at −80 °C. In this way, enzyme activity can be kept for at least 3 months. Enzyme activity was determined as described by Duan et al. (27). Microsomes (100 μg) were incubated with 10 μm of 14C-labeled substrates in the presence of 20 mm potassium phosphate (pH 7.4), 6.7 mm glucose 6-phosphate, 1 unit glucose-6-phosphate dehydrogenase, and 1 mm NADPH at 30 °C in a final volume of 50 μl. Reactions were initiated by the addition of microsomal protein followed by 15-min incubation and terminated by adding 5 μl of 6 n HCl. Metabolites were then converted to FAMEs as described below. Methylated metabolites were dissolved in 15 μl of hexane and then resolved by TLC. Radioactive signals were detected with an instant imager as described above.
Fatty Acid and Lipid Analyses
FAMEs of stigmas from petunia plants were prepared by placing the tissue in 1.5 ml of 2.5% H2SO4 (v/v) in methanol. After 1-h incubation at 80 °C, 1 ml of 0.9% NaCl and 1.5 ml of hexane were added. The upper phase was transferred to a glass tube, and then the lower phase was extracted with an additional 1.5 ml of hexane. The pooled organic phases containing FAMEs were evaporated under nitrogen gas. The FAMEs were dissolved in 50 μl of hexane and further treated with 50 μl of the silylating reagent N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% (v/v) trimethylchlorosilane (Supelco, Bellefonte, PA). The trimethylsilyl esters (TMSs) were dissolved in 200 μl of hexane, and a 1-μl sample was separated by GC on a 30-m × 0.25-mm Rtx-2330 capillary column (Restek, Bellefonte, PA). Chromatography was programmed with an initial temperature of 100 °C for 3 min, followed by a 20 °C/min ramp to 250 °C, and then held for 3 min. The eluted components were identified by their 70-eV electron-impact mass spectra scanning from 50 to 500 atomic mass units using a GCQ-coupled Polaris mass spectrometer (ThermoQuest, San Jose, CA).
For analysis of lipid species from transgenic plants, fresh stigmas (100 mg) were extracted by using the chloroform-methanol method (37). The extracted lipids were then dissolved in 1 ml of chloroform and separated by HPLC on an SP-silica column (Cobert, St. Louis, MO). The mobile phase consisted of hexane:isopropanol (9:1, v/v) for 5 min followed by hexane:isopropanol (3:1, v/v) for 10 min. A portion of each fraction was resolved by TLC on a silica gel 60 plate with a solvent system consisting of hexane:diethyl ether:formic acid (70:30:2, v/v) and visualized by iodine vapor. The remaining fraction was infused at 10 μl/min into the electrospray ionization interface of an LCQ ion trap mass spectrometer (ThermoQuest). The mass spectrometer was operated in positive ion mode with the source voltage set to 4.5 kV, a capillary voltage of 19 V, and temperature of 290 °C. In full MS mode, scans were collected between m/z values of 200 and 2000 to identify the molecular ion of individual fractions. In MS/MS mode, parent ions were fragmented with collision energy between 34 and 36%, and scanned out between m/z values of 200 and 1200.
Acyl-CoA Analysis
14C-Labeled acyl-CoA substrates were analyzed according to Hlousek-Radojcic et al. (38). The reactions were prepared as described under “Enzyme Assays” but stopped by the addition of two reaction volumes of 1-butanol:acetic acid, 5:2 (v/v). The phase was split by the addition of six reaction volumes of water, mixing well, and centrifugation. The upper butanol phase was collected and evaporated with a stream of nitrogen to near dryness before loading onto a silica gel 60 plate (EMD, Gibbstown, NJ) with a solvent system consisting of 1-butanol:acetic acid:water, 5:2:3 (v/v). The radioactive signals were detected with an instant imager as described above.
Acyl-CoAs were extracted from stigmas (∼60 mg) according to Larson and Graham (39) and analyzed using a 4000 Q-TRAP LC/MS/MS system (Applied Biosystems, Foster City, CA). The stigma acyl-CoAs were separated by HPLC on an Eclipse XDB-C18 column (3.0-mm × 100-mm with 3.5-μm particles, Agilent) fitted with a guard cartridge. Injection volume was 20 μl. The flow rate was set to 1 ml/min. The solvent system consisted of solvent A (acetonitrile:water, 10:90, v/v), solvent B (acetonitrile), and solvent C (water:acetonitrile:formic acid, 30:70:0.1, v/v). The gradient elution was initiated with 0% B and increased to 25% B/75% A in 5 min, and further to 50% B/50% A in 3 min and held for 2 min, then switched to 100% C in 2 min and held for 3 min, and finally changed to 100% A in 2 min and held for 5 min for equilibration.
Electrospray ionization-MS/MS spectrometry was performed in positive ion mode. Spray voltage was set to 5 kV, nebulizing gas (GS1) at 60 p.s.i., focusing gas (GS2) at 60 p.s.i., and curtain gas at 20 p.s.i. The source temperature was held at 750 °C. Declustering potential and collision energy were optimized on a compound-dependent basis.
The ω-hydroxyl acyl-CoAs were identified in multiple-reaction monitoring mode modified from Magnes et al. (40). In this mode, the precursor-product ion pairs of ω-hydroxyl acyl-CoAs were monitored by the addition of m/z 16 for the hydroxyl group to corresponding normal acyl-CoAs. For example, ω-hydroxyloleoyl-CoA was monitored by the precursor-product ion pair of m/z 1048.35 and 541.36, and ω-hydroxyllinoeoyl-CoA was monitored by the precursor-product ion pair of m/z 1046.33 and 539.34.
Sequence Analysis
Pairwise comparisons of amino acid sequences were performed using the MegAlign program (DNAStar Inc., Madison, WI). Multiple sequence alignments were done using ClustalX (41) and GeneDoc software6 or Vector NTI (Invitrogen). For phylogenetic analysis, an unrooted tree was developed using TreeView (42).
RESULTS AND DISCUSSION
Accumulation of ω-Hydroxy Fatty Acids in the Developing Stigma of P. hybrida
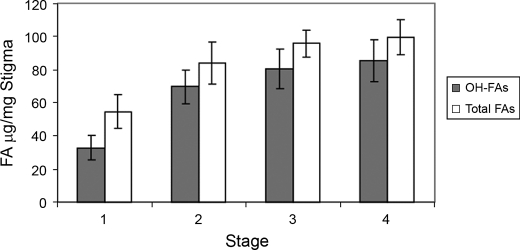
To generate a cDNA library representing genes for the biosynthesis of ω-hydroxy fatty acids in the stigma, we first determined the fatty acid composition in developing stigmas at four different stages by GC-MS analysis. As shown in Fig. 1, ω-hydroxy fatty acids represented the predominant fatty acid species in all four stages of stigma development. The amount of ω-hydroxy fatty acids in the stage 2 stigma nearly doubled relative to the stage 1 stigma. Beyond the early stages, the increase was less dramatic with stage 3 and stage 4 exhibiting only a 15 and 6% increase, respectively, over the preceding stage. These observations suggest that, in P. hybrida, stage 2 stigmas exhibit the fastest accumulation rate of ω-hydroxy fatty acids among the four stages of floral development. We therefore concluded that stage 2 was the most appropriate stage for isolation of mRNA for cDNA library construction.
FIGURE 1.
Accumulation of ω-hydroxy fatty acids (OH-FAs) in the different stage stigmas of floral buds of wild-type plants. Fatty acids were determined as FAME/TMS derivatives by GC-MS. Values are means with the error bar obtained from at least three independent samples.
Cloning of Putative Cytochrome P450s from the Petunia Stigma
An EST-based strategy was used to isolate P450 clones from the stage 2 stigma cDNA library. Forty-one of ∼10,000 ESTs were found to encode presumptive P450s. Based on their sequence identity, the P450 ESTs were classified into six groups, PH1–PH6, in which PH1 was represented by 30 ESTs; PH2, represented by six ESTs; and PH3–PH6, represented by one to two ESTs. Full-length cDNAs for each class were subsequently isolated from the cDNA library. In keeping with established nomenclature,7 we will hereafter refer to the deduced polypeptides, PH1–PH6, as CYP86A22, CYP714G3, CYP749B1, CYP94A13, CYP704A14, and CYP88C1, respectively. The six P450 clones all exhibit the conserved motifs characteristic of established P450 sequences, including the heme binding and oxygen activation motifs (supplemental Fig. 1). Among one another, however, they share relatively low amino acid sequence identity (11 - 33%), indicating that they belong to different P450 families. We further compared the six stigma clones to other P450 sequences (supplemental Fig. 2). The phylogenetic analysis reveals that CYP86A22, CYP94A13, and CYP704A14 are most closely related to established plant fatty acid ω-hydroxylases, whereas the remaining clones group with P450 enzymes likely involved in alternative pathways. CYP86A22 shares 70% amino acid sequence identity with CYP86A2 and CYP86A8, and CYP94A13 has 60% sequence identity with CYP94A1 and CYP94A2. CYP704A14 displays lower amino acid sequence identity (∼30%) with the CYP86A and CYP94A subfamily members, but higher identity (∼54%) with CYP704A subfamily members. Although the exact function of the CYP704 family members has not yet been clarified, as discussed above, both CYP86 and CYP94 families are documented fatty acid ω-hydroxylases involved in the synthesis of cutin and suberin polyesters (28–31). Thus, CYP86A22, CYP94A13, and CYP704A14 are all viable candidates for fatty acid ω-hydroxylases and were subjected to further characterization.
Because microsomal P450 hydroxylases require electron transfer partners for activity, we also cloned two putative NADPH:P450 reductases from the stigma cDNA library. Sequence analysis revealed that these cDNAs encode two closely related NADPH:P450 reductases, PR1 and PR2, with 66% amino acid sequence identity to each other (data not shown). Further sequence analysis showed that the PR1 and PR2 proteins share 72 and 68% amino acid identity with the A. thaliana P450 reductases ATR1 and ATR2, respectively (19, 20). These observations indicate that both PR1 and PR2 are candidates for NADPH:P450 reductases.
Expression Analysis of CYP86A22, CYP94A13, and CYP704A14
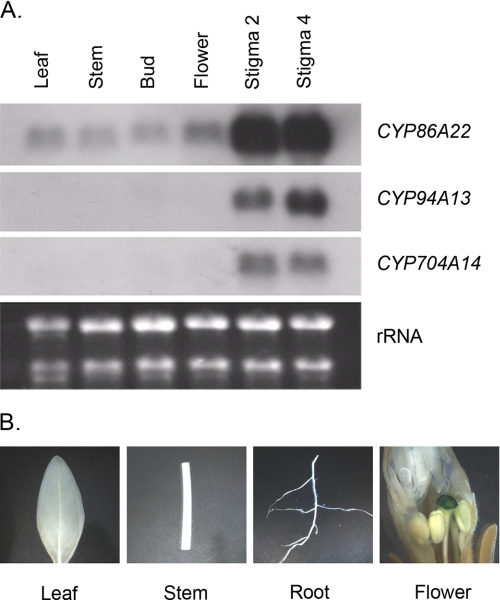
Because three P450 clones, namely CYP86A22, CYP94A13, and CYP704A14, fall into the plant fatty acid ω-hydroxylase group, we first examined their organ-specific mRNA expression. The Northern blot revealed that the CYP86A22 transcript is most abundant in the developing stigma of floral buds with virtually equivalent expression in both the stage 2 and stage 4 stigmas (Fig. 2A, upper panel). The transcript is also very weakly detected in the other tissues examined. Like CYP86A22, the transcripts of the CYP94A13 and CYP704A14 genes are almost exclusively restricted to the stigma (Fig. 2A, middle panels).
FIGURE 2.
Expression pattern of the putative fatty acid ω-hydroxylase genes in wild-type plants. A, Northern blot analyses. Total RNA (∼3 μg) from various tissues was hybridized with DIG-labeled probes derived from the P450 cDNAs. Stigma was removed from either bud or flower tissues used in this study. Stigma 2 and Stigma 4 represent the stage 2 and 4 stigma, respectively. An ethidium bromide stain of the gel served as a loading control. B, histochemical analyses of transgenic petunia plants containing the CYP86A22 promoter fused to the GUS reporter gene.
The CYP86A22 expression profile is supported by GUS reporter studies. In petunia transformed with the GUS reporter gene fused to the CYP86A22 promoter, a pattern of GUS expression similar to CYP86A22 transcript was observed. As shown in Fig. 2B, histochemical analysis revealed strong GUS activity in the stigma of transgenic petunia but not in other tissues such as leaves, stems, and roots. This result is essentially consistent with the Northern analysis, indicating that the CYP86A22 gene is predominantly expressed in the stigma of petunia.
CYP86A22 Encodes a Fatty Acid Hydroxylase Essential for the Production of ω-Hydroxy Fatty Acids in the Wet Stigma
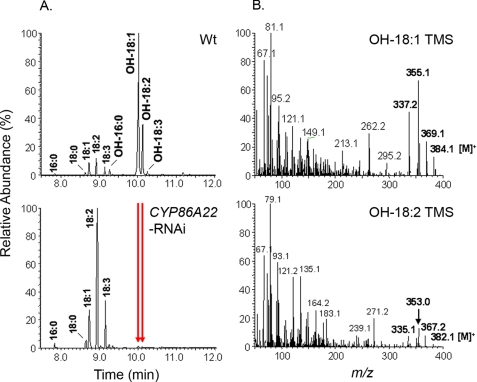
To probe the function of each P450, we generated RNAi constructs driven by the CYP86A22 promoter. The resulting RNAi constructs were introduced into the petunia plants. A total of 46 transgenic lines were obtained for CYP86A22-RNAi and 30 transgenic lines, respectively, for CYP94A13-RNAi and CYP704A14-RNAi. Fatty acid composition of the wild-type and the RNAi stigmas was determined by GC-MS analysis. As shown in Fig. 3A, fatty acid composition in the CYP86A22-RNAi stigma is dramatically altered. Compared with the wild-type stigma, which contains up to 96% ω-hydroxy fatty acids (Fig. 3A, upper panel), the ω-hydroxy fatty acid content in the typical CYP86A22-RNAi stigma is reduced to trace or non-detectable levels (Fig. 3A, lower panel). In contrast, no change in fatty acids was observed in CYP94A13- and CYP704A14-RNAi stigmas (data not shown). As shown in Fig. 3B, the mass spectra of ω-hydroxy fatty acid TMS derivatives of wild-type stigma are essentially identical to previous reports (10, 43), i.e. molecular ions [M]+ = 384 for OH-18:1 TMS and [M]+ = 382 for OH-18:2 TMS and their diagnostic fragment ions [M-15]+, [M-29]+, and [M-47]+. A comprehensive analysis of the stigma fatty acid composition of all CYP86A22-RNAi lines is presented in Table 1. The lines are grouped into six categories (Types I–VI) depending upon the amount of ω-hydroxy fatty acids measured in the stigmas. The majority of the CYP86A22-RNAi lines (Types V and VI) displayed significantly decreased levels of ω-hydroxy fatty acids in their stigmas relative to those of wild-type plants, suggesting that CYP86A22 encodes a major fatty acid ω-hydroxylase required for the biosynthesis of ω-hydroxy fatty acids in the petunia stigma.
FIGURE 3.
Altered fatty acid profile in the CYP86A22-RNAi stigma of transgenic petunia lines. A, GC-MS analysis of fatty acid composition of the wild-type stigma (upper panel) and the RNAi stigma (lower panel). The arrows indicate the corresponding ω-hydroxy fatty acids disappearing in the CYP86A22-RNAi stigma. B, the mass spectra of TMS derivatives of ω-hydroxyoleic acid (OH-18:1, upper panel) and ω-hydroxylinoleic acid (OH-18:2, lower panel) of wild-type stigma.
TABLE 1.
Distribution of ω-hydroxy fatty acids in the CYP86A22-RNAi stigmas of transgenic petunia lines
The lines are grouped into six types based on the percentage of ω-hydroxy fatty acids (OH-FAs) found in the total amount of fatty acids in the stigma. OH-FAs were determined as FAME/TMS derivatives by GC-MS.
| Type | I | II | III | IV | V | VI |
|---|---|---|---|---|---|---|
| OH-FAs (%) | 80–90 | 60–70 | 30–50 | 10–20 | 2–5 | 0–1 |
| No. of lines | 2 | 4 | 8 | 4 | 10 | 18 |
| Percentage | 4.3 | 8.7 | 17.4 | 8.7 | 21.7 | 39.1 |
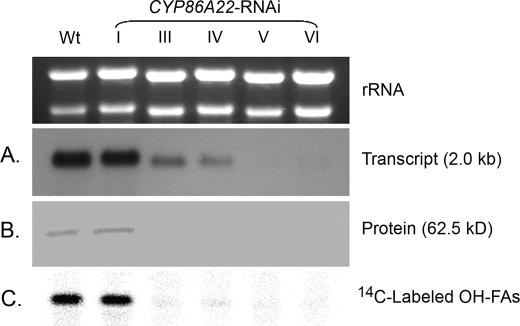
To further explore the correlation between ω-hydroxy fatty acid levels and CYP86A22, we examined CYP86A22 expression and activity in the different CYP86A22-RNAi stigma types (Fig. 4). Those RNAi stigmas containing high levels of ω-hydroxy fatty acids (Type I) also exhibit levels of CYP86A22 mRNA, protein, and activity that are commensurate with wild-type stigma. CYP86A22-RNAi stigmas with intermediate levels of ω-hydroxy fatty acids (Types III and IV) are characterized by reduced levels of CYP86A22 mRNA and greatly diminished levels of both CYP86A22 protein and ω-hydroxylase activity. Finally, CYP86A22-RNAi stigmas with trace or non-detectable levels of ω-hydroxy fatty acids (Types V and VI) also have no detectable CYP86A22 mRNA, protein, or hydroxylase activity. Collectively, these results strongly affirm that CYP86A22 is the fatty acid ω-hydroxylase responsible for ω-hydroxy fatty acid biosynthesis in the wild-type petunia stigma.
FIGURE 4.
Altered expression levels in the CYP86A22-RNAi stigma of transgenic petunia lines. A, relative levels of CYP86A22 mRNA were determined in stigmas exhibiting varying levels (I–VI) of ω-hydroxy fatty acids corresponding to Table 1. An ethidium bromide stain of the gel served as a loading control (above). B, microsomal proteins from the same stigma groups were subjected to Western analysis using the CYP86A22 antisera. C, conversion of exogenously supplied [1-14C] 18:1 fatty acid to ω-hydroxy 18:1 was evaluated in the different stigma types. FAMEs from these samples were separated on TLC plate, and radioactive signals were detected by an instant imager.
CYP86A22-derived ω-Hydroxy Fatty Acids Are Required for Estolide Biosynthesis
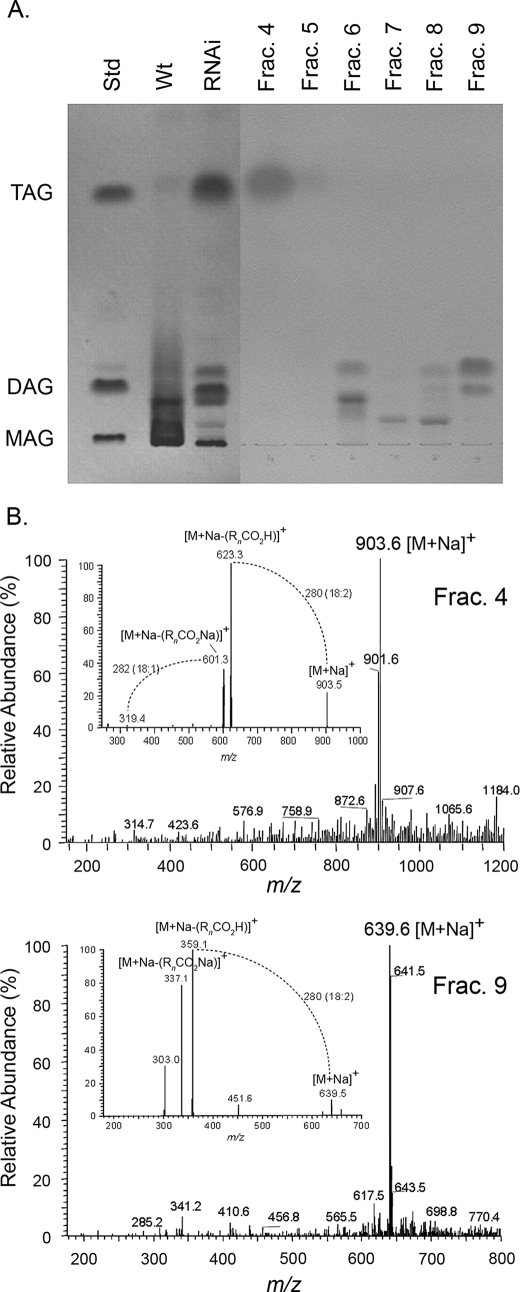
To determine if RNAi suppression of CYP86A22 has a global effect on lipid composition in petunia stigma, total lipids from either wild-type or CYP86A22-RNAi stigmas (Table 1, Type VI) were analyzed by both TLC and LC/MS. As shown in Fig. 5A, resolution of total lipids by TLC confirms that estolides are the predominant lipid forms in the wild-type stigma. Interestingly, in the TLC-resolved extract from the CYP86A22-RNAi stigma, estolides are essentially absent and have been replaced by TAGs and DAGs. No monoacylglycerols (MAGs), however, are found in the lipid extract. The total lipid extract from the CYP86A22-RNAi stigma was also fractionated by HPLC prior to TLC (Fractions 4–9). Selected fractions were subsequently analyzed by LC/MS, and their MS spectra were compared with authentic lipid standards. The most abundant ion of the Fraction 4 is m/z 903, reflecting [M + Na]+ of 18:2/18:2/18:1-TAG (Fig. 5B, upper panel). Its MS/MS yields fragment ions m/z 623 and m/z 601 that reflect the neutral loss of a fatty acid [M + Na − (RnCO2H)]+ and its sodium salt [M + Na − (RnCO2Na)]+, respectively, where n denotes the glycerol carbon to which the fatty acid is esterified (44, 45). As shown on the lower panel of Fig. 5B, the most abundant ion of Fraction 9 is m/z 639, reflecting [M + Na]+ of 18:2/18:2-DAG. Its MS/MS yields fragment ions m/z 359 and m/z 337 that also reflect the neutral loss of a fatty acid [M + Na − (RnCO2H)]+ and its sodium salt [M + Na − (RnCO2Na)]+, respectively. These data are consistent with the fatty acid analyses of RNAi stigma (Fig. 3A, lower panel), i.e. normal 18:2 fatty acid was the most abundant species in the RNAi stigma. Together, these findings suggest that the CYP86A22-derived ω-hydroxy fatty acids serve as monomers in the biosynthesis of estolides and that TAGs or DAGs serve to nucleate the formation of these polyesters.
FIGURE 5.
Altered lipid forms in the CYP86A22-RNAi stigma of transgenic petunia lines. A, TLC analysis of the wild-type and the RNAi stigma. Lipid standards used are as follows: 1,2,3-trilinoleoyl-sn-glycerol (TAG), 1,2-dioleoyl-sn-glycerol (DAG), and 1-oleoyl-rac-glycerol (MAG). Lipids were extracted from the wild-type stigma and the CYP86A22-RNAi type VI stigma and resolved by TLC. Lipid fractions from the RNAi type VI stigma were separated by HPLC with an SP-silica column. The major bands of fractions 6 and 8 are pigments. B, the full mass and MS/MS spectra (inset) of selected lipid fractions. The spectra of fraction 4 are shown in the upper panel and fraction 9 in the lower panel. [M + Na]+, sodium adduct of the molecular ion of TAG or DAG; [M + Na − (RnCO2H)]+ is loss of a fatty acid from [M + Na]+, and [M + Na − (RnCO2Na)]+ is loss of the sodium adduct of a fatty acid from [M + Na]+. n, the glycerol carbon to which the fatty acid is esterified.
Recombinant CYP86A22 Exhibits Fatty Acid ω-Hydroxylase Activity and Has a Substrate Preference for Acyl-CoAs
Although ω-hydroxy fatty acids are major components of stigmatic estolide, the enzymatic mechanisms underlying their synthesis are poorly characterized. Using RNAi technology, we have demonstrated that CYP86A22 is essential for the production of ω-hydroxy fatty acids in petunia stigma. To unambiguously confirm that CYP86A22 catalyzes the ω-hydroxylation of fatty acids, we infected Sf-9 insect cells with a recombinant baculovirus encoding CYP86A22, PR1, or PR2 and with a dual expression baculovirus encoding both CYP86A22 and PR1 or PR2. Microsomes were prepared, and ω-hydroxylase activity was examined using either 14C-labeled free fatty acids or the corresponding fatty acyl-CoAs as substrates. No ω-hydroxylase activity was detected in microsomes prepared from Sf-9 cells infected with wild-type baculovirus or recombinant virus encoding CYP86A22, PR1, or PR2 alone. Similarly, no ω-hydroxylase activity was observed when microsomes containing CYP86A22 alone were combined in vitro with microsomes containing PR1 or PR2 alone (data not shown). However, fatty acid ω-hydroxylase activity was observed in microsomes prepared from insect cells expressing the CYP86A22:PR1 dual recombinant baculovirus (Table 2). This activity was detected in the presence of an unsaturated fatty acid 18:1 but not with saturated free fatty acids such as 16:0 and 18:0. Surprisingly, significantly greater ω-hydroxylase activity was observed with both saturated and unsaturated fatty acyl-CoA substrates. Among the acyl-CoA substrates tested, the preference order in vitro was 16:0-CoA > 18:1-CoA > 18:0-CoA. Compared with the free fatty acid, activity was 3–20 times more with acyl-CoAs, suggesting that CYP86A22 has a preference for fatty acyl-CoA substrates. Similar activity was observed in microsomes prepared from Sf-9 cells expressing the CYP86A22:PR2 dual recombinant baculovirus (data not shown). Also, the lack of activity in microsomes containing CYP86A22 by itself implies that this enzyme cannot promiscuously use an endogenous Sf-9 reductase and instead requires a specific plant NADPH:P450 reductase for activity. This substrate specificity is clearly different from other ω-hydroxylases of the CYP86A or CYP94A subfamilies that are able to utilize various free fatty acids as substrates for cutin synthesis (26, 28, 29, 31).
TABLE 2.
Substrate specificity of CYP86A22 in the presence of the NADPH:P450 reductase PR1
Microsomes from insect cells infected with the CYP86A22:PR1 dual recombinant baculovirus were incubated with 10 μm 14C-labeled free fatty acids and acyl-CoAs at 30 °C for 15 min. Reaction products were analyzed as described under “Experimental Procedures.” Values are the mean of at least three independent assays ± S.E.
| Substrate | ω-Hydroxy fatty acid |
|---|---|
| pmol/min/mg protein | |
| 16:0 | NDa |
| 18:0 | ND |
| 18:1 | 6.7 ± 0.3 |
| 16:0-CoA | 110.0 ± 2.9 |
| 18:0-CoA | 19.3 ± 1.3 |
| 18:1-CoA | 23.3 ± 1.8 |
a ND, not detectable.
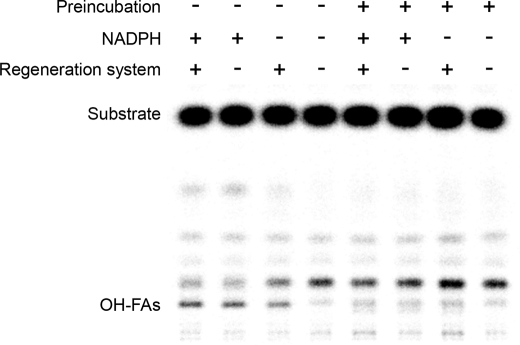
To examine whether acyl-CoA could be a direct substrate for CYP86A22, we performed preincubation assays in which the fatty-acyl moiety of the CoA substrate was allowed to incorporate into membrane lipids. As shown in Fig. 6, ω-hydroxy products were detected from microsomes directly incubated with acyl-CoA substrates in the presence of NADPH and/or the regeneration system. The conversion rates were 7.1%, 5.2%, and 4.2% in the presence of both NADPH and the regeneration system, NADPH alone, and the regeneration system alone, respectively. However, ω-hydroxy products were largely absent when the fatty-acyl moiety was first allowed to equilibrate with the microsomal membrane lipids. To ensure that this result was not a consequence of loss of CYP86A22 activity during preincubation, a separate experiment was conducted in which CYP86A22:PR1 microsomes alone were preincubated for various times prior to the addition of substrate (supplemental Fig. 3). The data unambiguously demonstrate that the CYP86A22 enzyme is stable during the 30-min preincubation. Moreover, the conversion rates of ω-hydroxy products at the 0-, 5-, 15-, and 30-min preincubation time points were essentially the same, i.e. 8.2%, 8.2%, 7.8%, and 8.0%, respectively. These observations indicate that the fatty-acyl moiety is unlikely first incorporated into a microsomal lipid intermediate that then serves as a substrate for CYP86A22. Rather, these results suggest that the acyl-CoAs are direct substrates for the hydroxylase.
FIGURE 6.
In vitro assays showing ω-hydroxy products. Microsomes from insect cells infected with the CYP86A22:PR1 dual recombinant baculovirus, without preincubation (lanes 1–4), were directly incubated with 10 μm [1-14C] 16:0-CoA at 30 °C for 15 min in the presence of both NADPH and the regeneration system (glucose 6-phosphate and glucose-6-phosphate dehydrogenase), NADPH alone, the regeneration system alone, or neither NADPH nor the regeneration system, respectively; microsomes were preincubated (lanes 5–8) with 10 μm [1-14C] 16:0-CoA at 30 °C for 30 min, followed by supplementation with both NADPH and the regeneration system, NADPH alone, the regeneration system alone, or neither NADPH nor the regeneration system, respectively, and incubated at 30 °C for an additional 15 min. Methylated products from these reactions were separated on a TLC plate, and radioactive signals were detected by an instant imager.
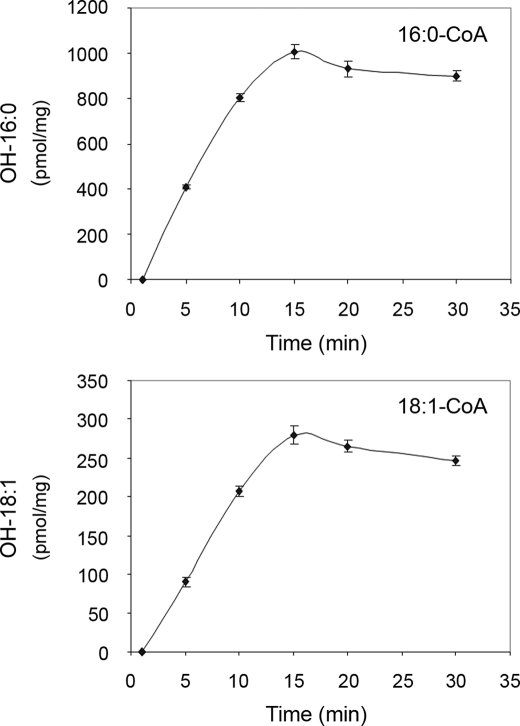
To further explore the enzymatic properties of CYP86A22, a time-course analysis of product formation was conducted. As shown in Fig. 7, the ω-hydroxy fatty acids steadily accumulated between 0 and 15 min, after which time ω-hydroxy product formation appeared to cease. A loss of ω-hydroxy product formation after 15 min might in part result from the competing utilization of the acyl-CoA substrate by endogenous enzyme systems such as desaturases and/or elongases.
FIGURE 7.
Progressive assays of CYP86A22. Time courses for 16:0-CoA (upper panel) and 18:1-CoA (lower panel). Microsomes from insect cells infected with the CYP86A22:PR1 dual recombinant baculovirus were incubated, respectively, with 10 μm [1-14C] 16:0-CoA and 18:1-CoA at 30 °C for various times ranging from 0 to 30 min in the presence of both NADPH and the regeneration system. Methylated ω-hydroxy products from these reactions were separated on TLC plate, and radioactive signals were detected by an instant imager.
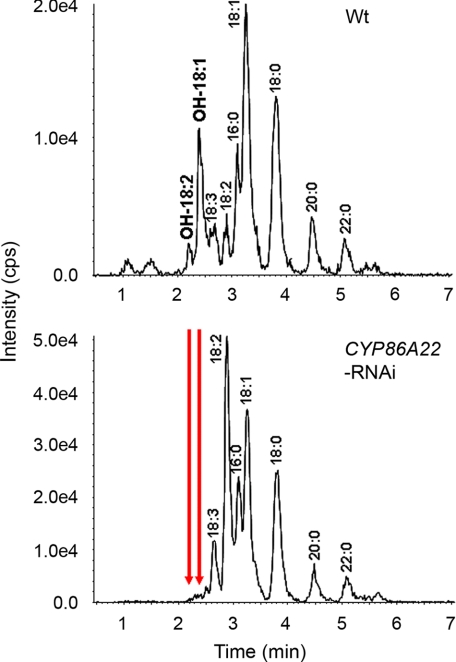
If the products of CYP86A22 are ω-hydroxy acyl-CoAs, we should be able to directly detect these products from petunia stigma instead of methylating the products of in vitro assays. To this end, we directly analyzed the ω-hydroxy acyl-CoA products of both wild-type and RNAi stigmas (Table 1, Type VI) using LC/MS/MS. As shown in Fig. 8, ω-hydroxy acyl-CoAs such as OH-18:1-CoA and OH-18:2-CoA were detected in the wild-type stigma, but not in the RNAi sigma, because the expression of CYP86A22 has been inhibited (Fig. 3A, lower panel, and Fig. 4). The same result was found using HPLC with fluorometric detection (data not shown). It is noted that relatively low levels of ω-hydroxy acyl-CoAs were observed compared with normal (non-ω-hydroxyl) acyl-CoAs, suggesting a much more efficient transesterification pathway involving estolide biosynthesis.
FIGURE 8.
In vivo analyses showing ω-hydroxy products. Acyl-CoAs were extracted from the wild-type stigma (upper panel) and the CYP86A22-RNAi type VI stigma (lower panel). The ω-hydroxy acyl-CoA products were directly analyzed by LC/MS/MS. The ω-hydroxy 18:1-CoA was monitored by the precursor-product ion pair of m/z 1048.35 and 541.36, and ω-hydroxy 18:2-CoA was monitored by the precursor-product ion pair of m/z 1046.33 and 539.34. The ion intensity was measured as counts per second (cps). The arrows indicate the corresponding ω-hydroxy acyl-CoAs disappearing in the CYP86A22-RNAi stigma.
As with the fatty acid analysis, we observed that normal linoleic acid is the major fatty acid species in the RNAi stigma (Fig. 3A, lower panel), while ω-hydroxyoleic acid is the predominant fatty acid in the wild-type stigma (Fig. 3A, upper panel). This observation implies that oleoyl-CoA may be a common substrate used for both desaturation and ω-hydroxylation in stigma, because the reduction of ω-hydroxyoleic acid in the RNAi stigma does not result in a corresponding increase in oleic acid.
From phylogenetic analyses (supplemental Fig. 2), it is clear that CYP86A22 displays ∼70% amino acid sequence identity with CYP86A2 and CYP86A8. Despite this conservation, these P450s exhibit differing substrate specificities. Moreover, no members of CYP86 and CYP94 family, two well studied families involved in fatty acid ω-hydroxylation, are known to use 18:0 as substrate. Further studies comparing CYP86A22 and these related P450s will likely provide insight as to the residues and domains responsible for these differing specificities.
In summary, we have identified CYP86A22 as the primary fatty acyl-CoA ω-hydroxylase responsible for the production of ω-hydroxy fatty acids in the stigma of P. hybrida. We further demonstrate that CYP86A22 derived ω-hydroxy fatty acids are essential for the formation of TAG-/DAG-based estolide polyesters that constitute the bulk of stigma exudate. Finally, we demonstrate that CYP86A22 has a clear substrate preference to both C16 and C18 saturated and unsaturated fatty acyl-CoAs. To date, no fatty acid ω-hydroxylase that uses an acyl-CoA substrate has been reported. This finding may provide new insight into the ω-hydroxylase family.
Supplementary Material
Acknowledgments
We thank Drs. John Ohlrogge and Mike Pollard for providing 14C-labeled ω-hydroxy 18:1 as standard and Dr. Jonathan Markham for his technical assistance in Q-TRAP LC/MS/MS.
This work was funded by Dow Chemical Co. (Midland, MI) and Dow AgroSciences LLC (Indianapolis, IN). The 4000 Q-TRAP system was supported by the National Science Foundation (Grant DBI-0521250).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) DQ099538, DQ099539, DQ099540, DQ099541, DQ099542, DQ099543, DQ099544, and DQ099545.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
K. Nicholas, H. Nicholas, and D. Deerfield (1997) GeneDoc, available on-line from the author.
D. R. Nelson (2009) The Cytochrome P450 Homepage.
- CYP
- cytochrome P450 monooxygenase
- RNAi
- RNA interference
- FAME
- fatty acid methyl ester
- TMS
- trimethylsilyl ester
- GC-MS
- gas chromatography-mass spectrometry
- LC/MS
- liquid chromatography/mass spectrometry
- MS/MS
- tandem mass spectrometry
- MAG
- monoacylglycerol
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- EST
- expressed sequence tag
- DIG
- digoxigenin
- PR
- P450 reductase
- RfB
- Gateway reading frame B
- GUS
- β-glucuronidase
- MES
- 4-morpholineethanesulfonic acid
- HPLC
- high performance liquid chromatography.
REFERENCES
- 1.Lord E. M., Russell S. D. (2002) Annu. Rev. Cell Dev. Biol. 18, 81–105 [DOI] [PubMed] [Google Scholar]
- 2.Edlund A. F., Swanson R., Preuss D. (2004) Plant Cell 16, (suppl.) S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konar R. N., Linskins H. F. (1966) Planta 71, 356–371 [DOI] [PubMed] [Google Scholar]
- 4.Martin F. W. (1969) Am. J. Bot. 56, 1023–1027 [Google Scholar]
- 5.Kandasamy M. K., Kristen U. (1987) Ann. Bot. 60, 427–437 [Google Scholar]
- 6.MacKenzie C. J., Yoo B. Y., Seabrook J. E. (1990) Am. J. Bot. 77, 1111–1124 [Google Scholar]
- 7.Matsuzaki T., Koiwai A., Kawashima N. (1983) Plant Cell Physiol. 24, 207–213 [Google Scholar]
- 8.Koiwai A., Matsuzaki T. (1988) Phytochemistry 27, 2827–2830 [Google Scholar]
- 9.Matsuzaki T., Koiwai A., Kawashima N. (1983) Agric. Biol. Chem. 47, 77–82 [Google Scholar]
- 10.Matsuzaki T., Koiwai A., Kubo S. (1986) Agric. Biol. Chem. 50, 1581–1587 [Google Scholar]
- 11.Konar R. N., Linskins H. F. (1966) Planta 71, 372–387 [DOI] [PubMed] [Google Scholar]
- 12.Cresti M., Keijzer C. J., Tiezzi A., Ciampolini F., Focardi S. (1986) Am. J. Bot. 73, 1713–1722 [Google Scholar]
- 13.Wolters-Arts M., Lush W. M., Mariani C. (1998) Nature 392, 818–821 [DOI] [PubMed] [Google Scholar]
- 14.Wolters-Arts M., Van der Weerd L., Van Aelst A., Van der Weerd J., Van As H., Mariani C. (2002) Plant Cell Environ. 25, 513–519 [Google Scholar]
- 15.Kolattukudy P. E. (2001) Adv. Biochem. Eng. Biotechnol. 71, 1–49 [DOI] [PubMed] [Google Scholar]
- 16.Nawrath C. (2002) in The Arabidopsis Book (Somerville C. R., Meyerowitz E. M. eds), The Biopolymers of Cutin and Suberin, pp. 1–14, American Society of Plant Biologists, Rockville, MD [Google Scholar]
- 17.Schuler M. A., Werck-Reichhart D. (2003) Annu. Rev. Plant Biol. 54, 629–667 [DOI] [PubMed] [Google Scholar]
- 18.Bernhardt R. (2006) J. Biotechnol. 124, 128–145 [DOI] [PubMed] [Google Scholar]
- 19.Urban P., Mignotte C., Kazmaier M., Delorme F., Pompon D. (1997) J. Biol. Chem. 272, 19176–19186 [DOI] [PubMed] [Google Scholar]
- 20.Mizutani M., Ohta D. (1998) Plant Physiol. 116, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson E. F., Palmer C. N., Griffin K. J., Hsu M. H. (1996) FASEB J. 10, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 22.Simpson A. E. (1997) Gen. Pharmacol. 28, 351–359 [DOI] [PubMed] [Google Scholar]
- 23.Okita R. T., Okita J. R. (2001) Curr. Drug Metab. 2, 265–281 [DOI] [PubMed] [Google Scholar]
- 24.Seghezzi W., Sanglard D., Fiechter A. (1991) Gene 106, 51–60 [DOI] [PubMed] [Google Scholar]
- 25.Zimmer T., Ohkuma M., Ohta A., Takagi M., Schunck W. H. (1996) Biochem. Biophys. Res. Commun. 224, 784–789 [DOI] [PubMed] [Google Scholar]
- 26.Benveniste I., Tijet N., Adas F., Philipps G., Salaün J. P., Durst F. (1998) Biochem. Biophys. Res. Commun. 243, 688–693 [DOI] [PubMed] [Google Scholar]
- 27.Duan H., Schuler M. A. (2005) Plant Physiol. 137, 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tijet N., Helvig C., Pinot F., Le Bouquin R., Lesot A., Durst F., Salaün J. P., Benveniste I. (1998) Biochem. J. 332, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Bouquin R., Skrabs M., Kahn R., Benveniste I., Salaün J. P., Schreiber L., Durst F., Pinot F. (2001) Eur. J. Biochem. 268, 3083–3090 [DOI] [PubMed] [Google Scholar]
- 30.Xiao F., Mark Goodwin S., Xiao Y., Sun Z., Baker D., Tang X., Jenks M. A., Zhou J. M. (2004) EMBO J. 23, 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellesen K., Durst F., Pinot F., Benveniste I., Nettesheim K., Wisman E., Steiner-Lange S., Saedler H., Yephremov A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupasinghe S. G., Duan H., Schuler M. A. (2007) Proteins 68, 279–293 [DOI] [PubMed] [Google Scholar]
- 33.King A., Nam J. W., Han J. X., Hilliard J., Jaworski J. G. (2007) Planta 226, 381–394 [DOI] [PubMed] [Google Scholar]
- 34.Horsch R., Fry J., Hoffmann N., Eichholtz D., Rogers S., Fraley R. (1985) Science 227, 1229–123117757866 [Google Scholar]
- 35.Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benveniste I., Salaün J. P., Simon A., Reichhart D., Durst F. (1982) Plant Physiol. 70, 122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christie W. W. (2003) in Lipid Analysis: Isolation, Separation, Identification and Structural Analysis of Lipids (Christie W. W. ed) pp. 91–135, The Oily Press, Bridgewater, UK [Google Scholar]
- 38.Hlousek-Radojcic A., Imai H., Jaworski J. G. (1995) Plant J. 8, 803–809 [Google Scholar]
- 39.Larson T. R., Graham I. A. (2001) Plant J. 25, 115–125 [DOI] [PubMed] [Google Scholar]
- 40.Magnes C., Sinner F. M., Regittnig W., Pieber T. R. (2005) Anal. Chem. 77, 2889–2894 [DOI] [PubMed] [Google Scholar]
- 41.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page R. D. (1996) Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 43.Kolattukudy P. E., Agrawal V. P. (1974) Lipids 9, 682–691 [Google Scholar]
- 44.Hsu F. F., Turk J. (1999) J. Am. Soc. Mass Spectrom. 10, 587–599 [DOI] [PubMed] [Google Scholar]
- 45.Holcapek M., Jandera P., Zderadicka P., Hrubá L. (2003) J. Chromatogr. A 1010, 195–215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.