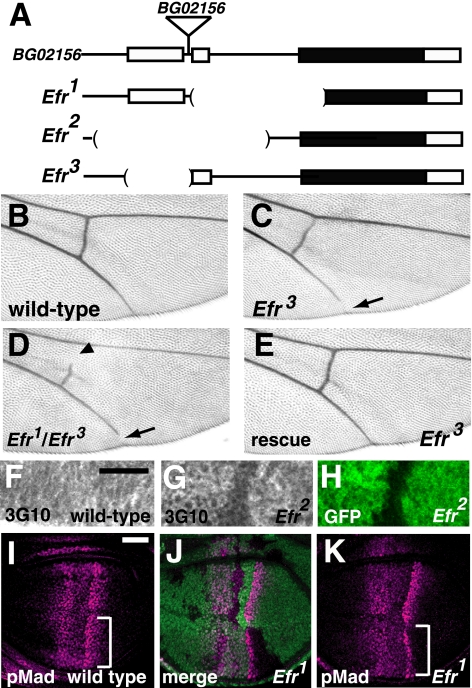
FIGURE 2.
Efr was involved in HS-GAG biosynthesis in vivo. A, genomic organization of the Efr locus. The exons of the Efr gene are shown as boxes, and the predicted coding regions are filled in black. The genomic regions deleted in the Efr1, Efr2, and Efr3 mutants are indicated by parentheses. In the Efr1 mutant, a ∼1.2-kb genomic region that includes a predicted initiation codon, was deleted. In the Efr2 mutant, a ∼1.5-kb genomic region that includes the first and second exons was deleted. In the Efr3 mutant, a ∼0.5-kb genomic region that includes the first exon was deleted. BG02156 was the original P-element insertion line, and a triangle indicates the position of the P-element insertion site. B–E, adult wings. B, a wild-type wing. C and D, a wing of Efr3/Efr3 (C) and a wing of Efr3/Efr3 (D), demonstrating the gap in the fifth longitudinal vein (arrow). In D, a gap in the posterior cross-vein was also observed (arrowhead). E, a wing of Efr3/Efr1 carrying a transgene of the Efr genome fragment. The wing vein gap was completely rescued. F–H, anti-heparan sulfate (3G10) staining of the late third instar wing discs. F, a wild-type wing disc. G and H, a chimeric wing disc, including Efr2/Efr2 cells, which are marked by the absence of GFP (H). Note that the intensity of anti-heparan sulfate staining was reduced in the mutant cells (G). I–K, late third instar wing discs stained with an anti-p-Mad antibody (magenta). I, a wild-type wing disc. J and K, a chimeric wing disc, including Efr1/Efr1 cells, which are marked by the absence of GFP (green in J). Anti-p-Mad antibody staining was decreased in Efr1 mutant cells (regions indicated by brackets in I compared with that of K). Bar, 25 μm.