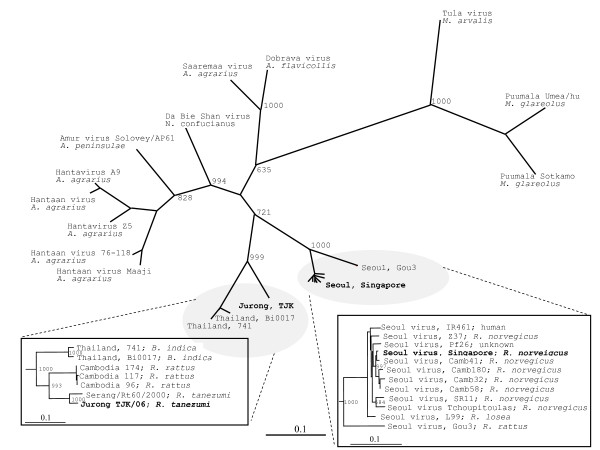
Figure 3.
Phylogenetic trees of hantavirus S sequences. Phylogenetic trees of hantaviruses based on the complete open reading frames (upper top figure) and partial S segment sequences (bottom left and right figures). In trees, the results after 1000 bootstrap replicates are indicated. (Upper top figure), Tula virus (Z69991); Puumala virus Umea/hu AY526219); Puumala Sotkamo (X61035); Dobrava-Belgrade virus (L41916); Dobrava/Saaremaa/160v (AJ009773); Da Bie Shan virus NC167 (AB027523); Amur virus Solovey/AP61/1999 (AB071183); Hantaan virus A9 (AF329390); Hantaan virus Hu (AB027111); Hantavirus Z5 (EF103195); Hantavirus 76-118 (AF321095); Seoul virus IR461 (AF329388); Hantaan Maaji (M14626); Jurong TJK/06 (this study); Thailand virus (Nakhon Ratchasima/Bi0017/2004) (AM397664); Thailand virus 741 (AB186420); Seoul virus Gou3 (AF184988); Seoul virus Singapore (this study). (Bottom left) Thailand virus (Nakhon Ratchasima/Bi0017/2004) (AM397664); Thailand virus 741 (AB186420); Cambodia 96 (AJ427512); Cambodia 174 (AJ427513); Cambodia 117 (AJ427511); Serang virus/Rt60/2000 (AM998808); Jurong TJK/06 (This study). (Bottom right) Seoul virus IR461 (AF329388); Seoul virus Z37 (AF187082); Seoul virus Pf26 (AY006465); Seoul virus Singapore (this study); Seoul virus Camb41 (AJ427501); Seoul virus Camb180 (AJ427506); Seoul virus Camb32 (AJ427508); Seoul virus Camb58 (AJ427510); Seoul virus SR11 (M34881); Seoul virus Tchoupitoulas (AF329389); Seoul virus L99 (AF288299); Seoul virus Gou3 (AF184988).