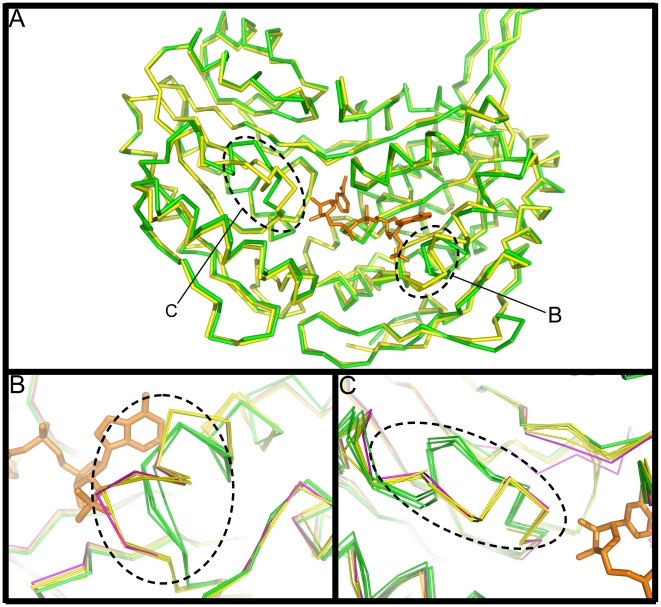
Figure 3. A single molecule superposition of E. coli SSADH and human SSADH.
A) Cα trace of monomer A of E. coli SSADH (green) superposed with the human SSADH molecule (PDB ID: 2w8r [26]: yellow: r.m.s.d. = 0.712 over 473 residues), with the NADP+ moiety (orange) from E. coli SSADH. Two structurally variable regions have been highlighted with dashed lines and labelled B–C. Figures B–C show Cα traces of all four E. coli SSADH monomers A–D (green) and 5 human SSADH monomers (open loop, PDB ID: 2w8o, 2w8p, 2w8q, 2w8r yellow; closed loop, PDB ID: 2w8n magenta)[26] superposed onto each other, only one NADP+ molecule (orange) from monomer A of E. coli SSADH is shown. B) Shows the region surrounding the 3 amino acid insertion (261RKN263) in human SSADH, which clashes with the 2'phosphate of NADP+. C) The loop motif connecting s2D and s3D in the catalytic domain, residues A379–G388 in E. coli SSADH and M432–G441 in human SSADH (r.m.s.d. of 3.4 Å over 10 residues). The E. coli SSADH loop is conserved throughout the ALDH family and is stabilised by 7 hydrogen bonds. The novel loop in human SSADH is stabilised by only 3 hydrogen bonds, furthermore this same loop in the reduced wild type human SSADH (PDB ID: 2w8o)[26] is highly flexible and could not be determined using X-ray crystallography.