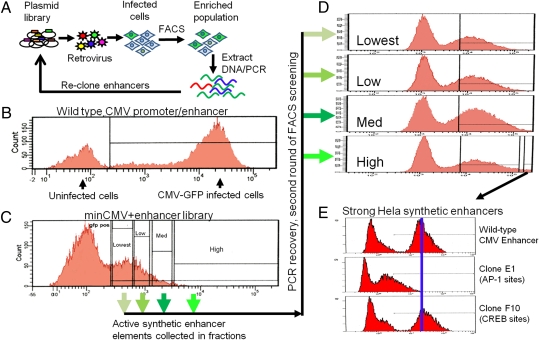
Fig. 2.
Screening synthetic enhancers for transcriptional activation. (A) The synthetic enhancer plasmid library is packaged as retroviruses and infected into tissue culture cells at an MOI of 0.1, puromycin-selected, and screened by FACS for cells expressing the desired level of GFP. After sorting and genomic DNA preparation, the synthetic enhancer inserts are PCR-amplified and recloned for subsequent rounds of screening. (B). Histogram of GFP fluorescence of HeLa cells expressing GFP under the control of the WT CMV promoter and enhancer. (C) Histogram of GFP fluorescence of HeLa cells expressing GFP driven by a minimal CMV promoter with the synthetic enhancer library cloned upstream. Populations indicated on the FACS histogram were separately sorted, and synthetic enhancers present were recloned and reinfected into HeLa cells. (D) Histograms of GFP fluorescence for cells infected in the second round of screening, with arrows indicating their parental population of cells from the first round of screening. (E) After two rounds of screening, clones were tested individually for transcriptional activity. A clone representing an enhancer with relatively weak activity (clone E1) and one with very potent activity (clone F10) are compared with the WT CMV promoter/enhancer. The predicted transcription factor binding sites for each synthetic enhancer are indicated.