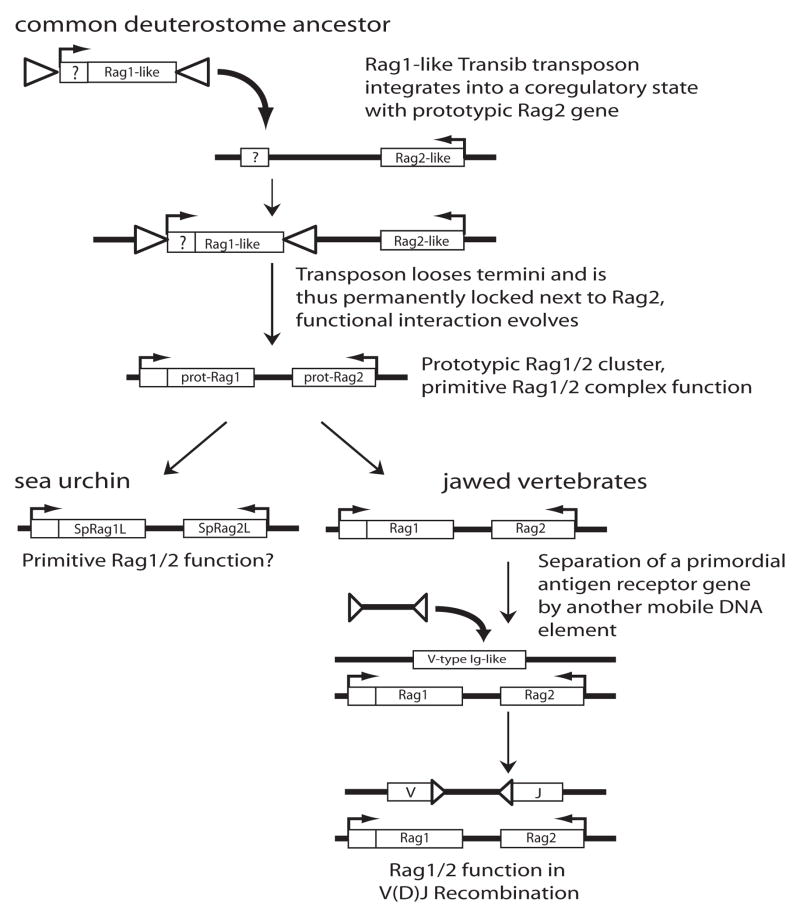
Figure 2. Current model of the origins of the Rag1/Rag2 gene cluster.
This is a schematic representation of the “Rag1-only” transposon concept, and also illustrates the “two step” concept of the evolution of V(D)J recombination (see section Open Questions). Coding regions are drawn as open boxes with the name of the gene inside, RSS-like TIRs and RSSs of flanking mobile elements as triangles, and directions of transcription are shown by arrows. In a common ancestor of all living deuterostomes, a Rag1-like transposable element integrates next to a Rag2-like gene. The box with “?” indicates that the N-terminus of Rag1 may have either been part of the original transposon or of a gene into which it integrated. The TIRs flanking the transposon are lost, and a functional interaction of the prototypical Rag1 and Rag2 (prot-Rag1 and prot-Rag2) proteins emerged, serving an unknown primitive function. As the lineages that led to sea urchins and jawed vertebrates diverged, the sea urchin gene cluster (SpRag1L/SpRag2L) may have retained this original function, while the vertebrate genes (Rag1/Rag2) evolved into the V(D)J recombinase. The disruption of a V-type Ig-like receptor gene by an RSS-flanked mobile element, an event that is predicted to have occurred in a common ancestor of all jawed vertebrates, represents a critical step in this process.