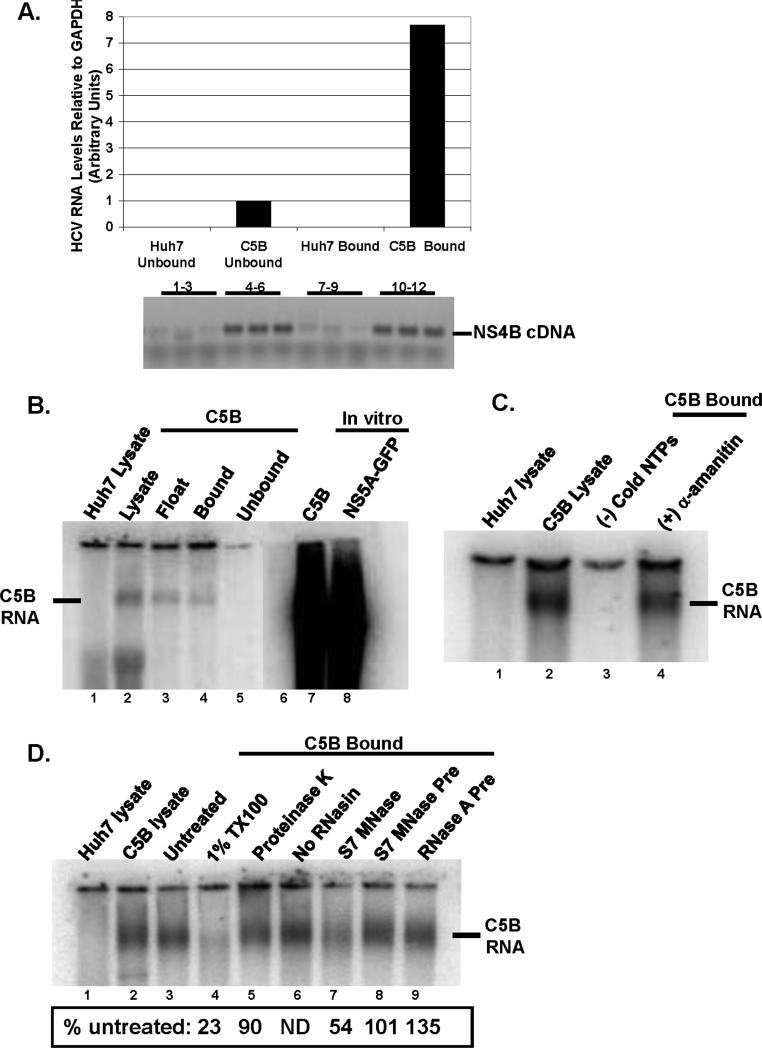
FIG.2.
A. RNA was extracted from bound and unbound fractions from Huh7 and C5B cells. The relative amount of RNA was determined using real-time PCR and was normalized to GAPDH levels. cDNA products were also run on a 1% agarose gel: lanes 1-3 (Huh7 unbound) and 7-9 (Huh7 bound) show no product whereas lanes 4-6 (C5B unbound) and 10-12 (C5B bound) display HCV-specific cDNA. B. The samples from different steps in the isolation procedure were tested for in-vitro RdRp activity in the absence of exogenous HCV RNA template. In-vitro transcribed and radiolabeled C5B (lane 7) and NS5AGFP (lane 8) replicon RNAs were included as markers. Notice the presence of a product in C5B lysate (lane 2) and C5B bound (lane 4). No such product is present in Huh7 lysate (lane 1). C. The bound fractions were tested for RdRp activity in the absence of cold NTPs (lane 3) or in the presence of an inhibitor of host RNA polymerase II, α-amanitin (lane 4). Notice that no RNA was detectable when RdRp assay was performed in the absence of cold NTPs (lane 3), but α-amanitin coupled with actinomycin D had little effect on RdRp activity (compare lane 4 to lane 1). D. The samples were tested for in-vitro RdRp activity in the presence of 1% TX100 (lane 4), 0.5 U/ml S7 MNase (lane 7), 1mg/ml proteinase K (lane 5) or preincubated with 0.5 U/ml RNases (lanes 8 and 9). In lane 6, RdRp activity was tested in the absence of RNase inhibitor, RNasin. The reaction mixtures were processed as in FIG. 2 B & C. Panels A through D are representative results from at least two independent experiments. No RNAsin (Fig. 4D, lane 6) was done only once.