Abstract
Eukaryotic cells maintain proteostasis by quality control (QC) degradation. These pathways can specifically target a wide variety of distinct misfolded proteins, and so are important for management of cellular stress. Although a number of conserved QC pathways have been described in yeast, the E3 ligases responsible for cytoplasmic QC are unknown. We now show that Ubr1 and San1 mediate chaperone-dependent ubiquitination of numerous misfolded cytoplasmic proteins. This action of Ubr1 is distinct from its role in the “N-end rule.” In this capacity, Ubr1 functions to protect cells from proteotoxic stresses. Our phenotypic and biochemical studies of Ubr1 and San1 indicate that two strategies are employed for cytoplasmic QC: chaperone-assisted ubiquitination by Ubr1 and chaperone-dependent delivery to nuclear San1. The broad conservation of Ubr ligases and the relevant chaperones indicates that these mechanisms will be important in understanding both basic and biomedical aspects of cellular proteostasis.
Keywords: chaperone, proteostasis, misfolding
Protein quality control (QC) functions to ensure that damaged and misfolded proteins are maintained at acceptable levels to limit their stress-causing, or proteotoxic, effects. One strategy of protein QC is the selective degradation of misfolded proteins. For degradative QC pathways to be effective, they must be specific for aberrant proteins; sufficiently general to recognize selectively common structural hallmarks shared by numerous unrelated proteins; and physiologically important, better allowing the cell to survive proteotoxic stress. Because protein QC underlies many pressing maladies, such as parkinsonism, cystic fibrosis, and aging, discovery of the rules of substrate selectivity and destruction is a key step in understanding these conditions and designing appropriate therapeutical interventions to combat them.
In eukaryotes, the ubiquitin proteasome system is employed in the selective degradation of many proteins (1). A substrate protein is marked for degradation by assembly of a polyubiquitin chain, initiated by covalent addition of the small (7.6 kDa) protein ubiquitin to a lysine in an isopeptide bond, followed by iterative addition of the next ubiquitin to the previously added one to create a polyubiquitin chain that is uniquely recognized by the 26S proteasome. Protein ubiquitination is catalyzed by a three-enzyme cascade. The single E1 ubiquitin-activating enzyme hydrolyzes ATP to acquire ubiquitin in labile thioester linkage, which is then transferred in thioester linkage to one of a small group of E2s or ubiquitin-conjugating enzymes (UBCs). E2-bound ubiquitin is finally transferred to an isopeptide linkage on the target protein or the growing polyubiquitin chain by the action of the E3 ubiquitin ligase. It is the E3 ubiquitin ligase that determines the specificity of a given ubiquitination process; identifying and understanding the E3s involved in a degradative pathway are thus key parts of understanding the mechanisms of substrate selection and modification.
E3s for several QC pathways have been discovered and include the endoplasmic reticulum-associated ligases Hrd1 and Doa10 involved in endoplasmic reticulum-associated degradation (ERAD) and the San1 ubiquitin ligase that mediates destruction of misfolded nuclear proteins (2–4). The mechanism used by the QC ligases to detect misfolded substrates can vary, with some employing chaperones (5) and others not. The details of substrate recognition are key to understanding the envelope of structures subject to destruction by a given pathway.
So far, no widely conserved ubiquitination pathway has been described for cytoplasmic QC. Metazoans express the CHIP ubiquitin ligase that mediates cytoplasmic QC, using Hsp70 chaperones to detect misfolded proteins (6, 7). However, CHIP is not conserved in all eukaryotes. For example, no CHIP is encoded in yeast. Nevertheless, chaperone-dependent ubiquitination of misfolded proteins has been observed in yeast (8), indicating that previously undescribed, and probably highly conserved, cytoplasmic QC pathways remain to be discovered.
To that end, we have investigated the E3 ligases involved in ubiquitination of misfolded cytoplasmic proteins in yeast. We have discovered that two E3 ligases collaborate in ubiquitination of a diverse set of misfolded proteins, including full-length substrates with point mutations and truncated proteins. The two E3s are the nuclear E3 San1 (9, 10) and Ubr1, best known in the “N-end rule” pathway (11, 12). A variety of misfolded substrates undergo selective chaperone-dependent ubiquitination by either ligase. In this function, Ubr1 and San1 appear to function independently. In vitro experiments indicate that the Ubr1 ligase directly employs chaperones in substrate ubiquitination, whereas the San1 E3 may require chaperones for delivery to the nucleus. Our phenotypic studies show that Ubr1 had the principal role in mediating cytoplasmic proteotoxic stress imposed by model substrates or chemical stressors. This QC function of Ubr1 was independent of its function in the well-described N-end rule, and so represents a previously undescribed physiologically important role for this molecule. Our demonstration of parallel pathways indicates the importance and complexity of cytoplasmic proteostasis. Understanding them will provide unique opportunities for management of damaged proteins in the clinical setting.
Results and Discussion
We launched a genetic study of CPY‡, a model cytoplasmic substrate derived from the misfolded vacuolar protease CPY*, in which the signal sequence is removed to restrict it to the cytosol (13). CPY‡ was fused to GFP, yielding the optically detectable misfolded cytoplasmic protein CPY‡-GFP (13, 14) (Fig. S1A). We expressed CPY‡-GFP from the constitutive pTDH3 promoter and confirmed that the strongly expressed protein showed similar degradative behavior to that previously reported for this substrate (Figs. S1 and S2): CPY‡-GFP degradation was mediated by the ubiquitin proteasome system and required Hsp70. In addition, we tested other chaperones and cochaperones and observed that Ydj1 (Hsp40) and Sse1 (Hsp110) were required, with Sse1 playing a major role in substrate ubiquitination, whereas Hsp104 and Sti1 did not play a role.
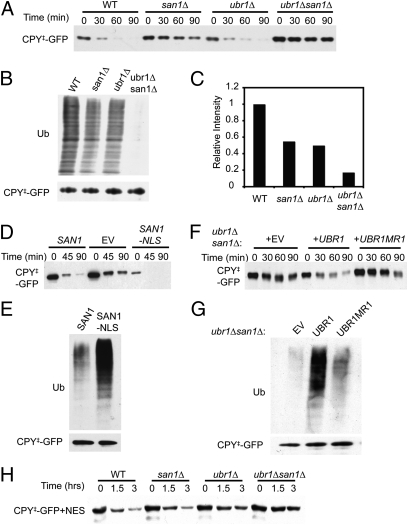
To find the relevant ligases, we screened an inclusive collection of yeast nulls in genes relevant to the ubiquitin pathway provided by the Hochstrasser laboratory (Yale Univ., New Haven, CT). A single genomically integrated CPY‡-GFP expression plasmid was introduced into each null strain by array mating (15), and CPY‡-GFP stability was assayed by cycloheximide chase and flow cytometry. Surprisingly, the san1Δ null stabilized CPY‡-GFP, despite the San1 E3 being involved in nuclear QC (9), showing stabilization (Fig. 1A) and decreased ubiquitination of CPY‡-GFP (Fig. 1 B and C). We tested if San1 could function in the cytoplasm to ubiquitinate CPY‡-GFP with San1 missing its nuclear localization signal (NLS) (9). San1-NLS was also fully competent for CPY‡-GFP degradation and ubiquitination (Fig. 1 D and E).
Fig. 1.
Ubr1 and San1 mediate CPY‡-GFP degradation and ubiquitination. (A) Cycloheximide chase of CPY‡-GFP in the suite of WT, san1Δ, ubr1Δ, and ubr1Δsan1Δ strains. Incubation time following cycloheximide addition is indicated in minutes. Anti-GFP immunoblotting. (B) CPY‡-GFP ubiquitination assayed in the suite of strains in A and IP with anti-GFP followed by immunoblotting for ubiquitin (Ub) or GFP. (C) Ubiquitination immunoblotting intensities normalized to the total precipitated CPY‡-GFP for each assay, as determined using ImageQuant TL. Results are graphed as a fraction of WT, which was set to 1.0. (D) Cycloheximide chase of CPY‡-GFP in san1Δ background with either SAN1, empty vector (EV), or SAN1-NLS expressed from plasmids. (E) Ubiquitination of CPY‡-GFP in the SAN1 or SAN1-NLS strain in D. (F) Cycloheximide chase of CPY‡-GFP from ubr1Δsan1Δ strains expressing UBR1 or the UBR1MR1 ring mutant compared with the EV strain. (G) Ubiquitination of CPY‡-GFP from the Ubr1-expressing strains in F. (H) Cycloheximide chase of cytoplasm-restricted CPY‡-GFP+NES in the strains used in A.
In the san1Δ null, there remained measurable CPY‡-GFP degradation and ubiquitination. To discover the remaining E3, we again employed the null collection, this time crossing a san1Δ strain expressing CPY‡-GFP to each null and selecting for haploids that bore the CPY‡-GFP plasmid, the san1Δ null, and each test null. The UBR1 gene accounted for the residual degradation (11, 16), and the ubr1Δsan1Δ double null showed nearly complete stabilization of the test substrate. We turned our attention to evaluating the participation of these two ligases in the destruction of misfolded cytoplasmic proteins.
A suite of strains, including WT, san1Δ, ubr1Δ, and san1Δubr1Δ, was prepared to assess the contribution of each ligase in cytoplasmic QC directly. The stability of CPY‡-GFP was affected by either single null, with san1Δ showing the larger effect (Fig. 1A). Each null decreased CPY‡-GFP ubiquitination to a similar extent, although the san1Δubr1Δ showed a stronger additive decrement in ubiquitination (Fig. 1 B and C). By comparing the two san1Δ lanes, it is clear that the Ubr1 ligase alone could mediate CPY‡-GFP ubiquitination. This was confirmed by adding a Ubr1-expressing plasmid to the san1Δubr1Δ double null. Ubr1 enhanced degradation and ubiquitination of CPY‡-GFP (Fig. 1 F and G), whereas the nonfunctional C1220S RING mutant of Ubr1 did not (UBR1MR1 in Fig. 1 F and G), indicating that Ubr1 ubiquitin ligase activity of the protein was required for this effect. Similarly, overexpression of Ubr1 caused increased degradation of CPY‡-GFP (Fig. S1E). To elucidate the roles of Ubr1 and San1 in degradation of the reporter, a nuclear export signal (NES) was placed at the C terminus of CPY‡-GFP (CPY‡-GFP+NES). Degradation of this cytoplasmically restricted substrate was now only dependent on Ubr1. The san1Δ null had no effect on degradation of the cytoplasmically restricted substrate, either alone or in combination with ubr1Δ (Fig. 1H).
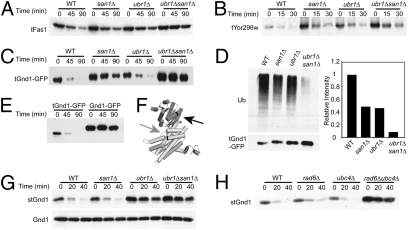
The generality of Ubr1 and San1 in cytoplasmic QC was revealed in a dosage suppression screen. We screened a yeast high-copy genomic library (2μ plasmids, ∼20 copies per cell) for plasmids that inhibited CPY‡-GFP degradation in a WT strain, using colony GFP fluorescence (17) and cycloheximide chase. There was a striking uniformity in the resulting coding regions. Each stabilizing plasmid included a truncated coding region that expressed a C-terminally truncated protein. In the cases examined, the truncated coding regions were verified to cause CPY‡-GFP stabilization. None of the candidate coding regions had any functional connection to protein degradation or misfolded proteins. Because truncations frequently result in proteins that fold incorrectly, we posited that the plasmids encode competing QC substrates. We tested this possibility with three truncated coding regions from the screen (% indicates the fraction of the coding region present)—FAS1 (48%), YOR296w (39%), and GND1 (75%)—encoding the truncated proteins tFas1, tYor296w, and tGnd1, respectively. FAS1 encodes the cytoplasmic multidomain fatty acid synthetase, YOR296w encodes an unknown protein predicted to reside in the cytosol, and GND1 encodes cytoplasmic phosphogluconate dehydrogenase. Epitope-tagged versions of each truncated protein were degraded in a Ubr1- and San1- dependent manner, and the three showed a range of degradation rates. Each truncated protein was stabilized by each null and strongly stabilized in the ubr1Δsan1Δ double null (Fig. 2 A and B). To study tGnd1 in more depth, it was expressed from a plasmid that added 3HA tags to the N terminus and GFP to the C terminus, producing 3HA-tGnd1-GFP, which was similarly degraded and ubiquitinated (Fig. 2 C and D). Importantly, the addition of GFP to tGnd1 created a full-length fusion protein with a folded C-terminal domain. Thus, the pathway defined by the CPY‡-GFP reporter is involved in the degradation of a variety of unrelated cytoplasmic proteins, including both truncated and misfolded full-length proteins.
Fig. 2.
Ubr1 and San1 mediate the degradation of multiple cytoplasmic QC substrates. Cycloheximide chase and immunoblot of HA-tagged truncated Fas1 (48% total, tFas1) (A), Yor296w (39% total, tYor296w) (B), and Gnd1 (75% total, tGnd1-GFP) (C) in the indicated strains. (D) San1 and Ubr1 dependence of tGnd1-GFP ubiquitination measured by anti-GFP IP, followed by immunoblotting with anti-GFP or anti-ubiquitin (Ub) as indicated (Left). (Right) Ubiquitination immunoblotting intensities were normalized to the total precipitated tGnd1-GFP for each strain, as determined using ImageQuant TL. Results are graphed as a fraction of WT, which was set to 1.0. (E) Cycloheximide chase and anti-HA immunoblotting of 3HA-tGnd1-GFP or full-length Gnd1 fused to GFP, 3HA-Gnd1-GFP. (F) Crystal structure of full-length yeast Gnd1 created using PyMOL (19). The black arrow indicates the stGnd1 truncation point. The gray arrow indicates the tGnd1 truncation point. (G) Cycloheximide chase of 3HA-stGnd1 (Upper) or 3HA-full-length Gnd1 (Lower) in the indicated strains. Detection was with anti-HA antibodies. (H) Cycloheximide chase of stGnd1 in E2 UBC strains rad6Δ and ubc4Δ as indicated.
QC pathways selectively degrade misfolded proteins, sparing the fully folded forms (e.g., 9). We evaluated this specificity with Gnd1. The original CPY‡-GFP reporter cannot be used in this test, because secretory proteins such as BSA (7) and nonmutant CPY-GFP do not correctly fold in the cytosol. Conversely, the full-length cytoplasmic Gnd1 is normally folded in the cytosol. 3HA-Gnd1-GFP, with full-length normal Gnd1 in the fusion, was quite stable in cells compared with the identically expressed 3HA-tGnd1-GFP (Fig. 2E). QC pathways are selectively able to degrade a large variety of misfolded proteins. This flexibility includes diverse substrates, or distinct misfolded versions of the same protein, as seen with the many unstable “type II” CFTR mutants (18). As a separate test of the generality of this cytoplasmic QC pathway, we used the x-ray structure of yeast Gnd1 protein to generate a unique QC substrate (19). The original tGnd1 substrate is truncated in the middle of the second folded domain (Fig. 2F, gray arrow; residues 1–368, 75%). The first domain is a widely observed structure that allows NADP+ binding with a Rossman fold. We tested a version of Gnd1p with a truncation in the first domain (Fig. 2F, black arrow; residues 1–150, 30%), calling the substrate stGnd1 (short truncated Gnd1). Indeed, 3HA-stGnd1 was rapidly degraded (Fig. 2G), showing a 10-min half-life in the presence of Ubr1 and strong stabilization in the ubr1Δ null. Interestingly, this substrate was entirely dependent on Ubr1; the presence of san1Δ alone or in combination with ubr1Δ had no effect on the stability of stGnd1. Identically tagged full-length Gnd1 was completely stable (Fig. 2G, Bottom). Thus, Ubr-mediated QC is specific for misfolded proteins but is broadly inclusive in selection of substrates.
We employed the Ubr1 specificity of stGnd1 degradation to evaluate the E2 enzymes involved in Ubr1-mediated QC. As observed in other actions of this E3 (20), both Rad6/Ubc2 and Ubc4 could mediate degradation and the double E2 null phenocopied the ubr1Δ strain (Fig. 2H).
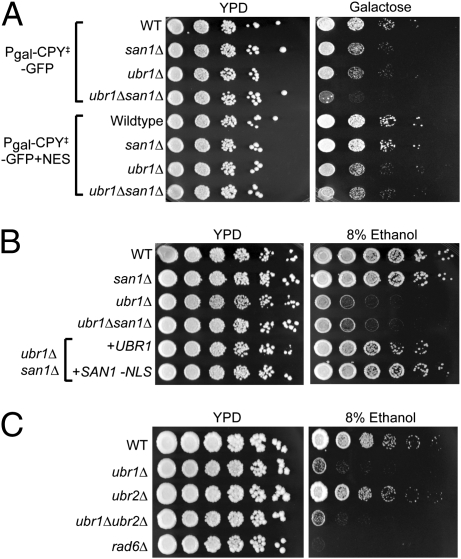
These studies show that both Ubr1 and San1 are required for degradation of a wide variety of misfolded proteins that originate in the cytoplasm. We next tested each ligase for roles in management of proteotoxic stress. We initially tested stress caused by overexpression of CPY‡-GFP. Although this protein can be expressed in any of the nulls without consequence from the TDH3 promoter in glucose medium, expression of CPY‡-GFP from the stronger galactose promoter caused growth sensitivity that was exacerbated by either the ubr1Δ or the san1Δ and greatly worsened in the double null (Fig. 3A, Top four rows). Thus, as expected from the CPY‡-GFP degradation data, both Ubr1 and San1 can lessen the stress caused by strong expression of this model substrate. Identical expression of the cytoplasm-restricted CPY‡-GFP+NES resulted in stress that was only affected by ubr1Δ; the presence of san1Δ alone or in combination with ubr1Δ had no additional effect on growth (Fig. 3A, Bottom four rows). This result highlights Ubr1’s direct role in survival of cytoplasmic proteotoxic stress.
Fig. 3.
Involvement of Ubr1 and San1 in managing proteotoxic stresses. (A) Cultures of strains expressing CPY‡-GFP (Top four rows) or cytoplasmic CPY‡-GFP +NES (Bottom four rows) under the control of the galactose promoter were spotted in 10-fold dilutions onto plates with either dextrose [yeast peptone dextrose (YPD)] or galactose as the carbon source and grown at 23°C. (B) Test of ethanol stress. WT, san1Δ, ubr1Δ, or ubr1Δsan1Δ null strains were spotted onto media containing 8% (vol/vol) ethanol in 5-fold serial dilutions and grown at 23°C as indicated. (Bottom two rows) Effect of expressing UBR1 or cytoplasmic SAN1-NLS in the ubr1Δsan1Δ strains is shown. (C) Same test of effect of 8% (vol/vol) ethanol on ubr1Δ, ubr2Δ, ubr1Δubr2Δ, or rad6Δ as indicated.
We tested a variety of growth conditions to evaluate physiological roles for these ligases, including ethanol, which has been employed as a proteotoxic stress (21). We found that 8% (vol/vol) ethanol in solid medium caused a significant cold-sensitive growth phenotype in the ubr1Δ null. The ubr1Δ strains showed a strong growth defect on the ethanol plates at 23°C (∼600-fold; Fig. 3B). In contrast, the san1Δ null showed no defect of growth, nor did it enhance the sensitivity of the ubr1Δ (Fig. 3B, san1Δ vs. WT and san1Δubr1Δ vs. ubr1Δ). Thus, full-length San1 was not involved in this ubr1Δ-sensitive stress. However, the growth phenotype of the ubr1Δsan1Δ double null was suppressed by cytoplasmic SAN1-NLS expressed in single copy from its native promoter (Fig. 3B, +SAN1-NLS). This strongly implied that the ethanol stress imposed in this test occurred in the cytosol and that it was remediated by destruction of misfolded proteins, either by normally present Ubr1 or by the cytoplasmic -NLS version of San1.
The ethyl alcohol (EtOH) growth phenotype was also observed in the absence of the E2 Rad6 (Fig. 3C, rad6Δ), which is implicated in many actions of Ubr1, including QC (Fig. 2G). The decreased survival in rad6Δ was more pronounced than in ubr1Δ, which may be attributable to Rad6 having numerous roles in cell function. Alternatively, the Ubr1 homolog Ubr2 (∼50% identical to Ubr1) also uses Rad6 (22) and could also have been involved in cytoplasmic QC. However, the ubr2Δ null showed no growth defect in the 8% (vol/vol) EtOH test (Fig. 3C), nor did the presence of the ubr2Δ null make the ubr1Δ sensitivity any greater (ubr1Δubr2Δ). Furthermore, the ubr2Δ null had no effect on degradation of our substrates, and overexpression of Ubr2 did not increase the degradation rate of any tested QC substrates. Thus, QC function is unique to the Ubr1 isozyme.
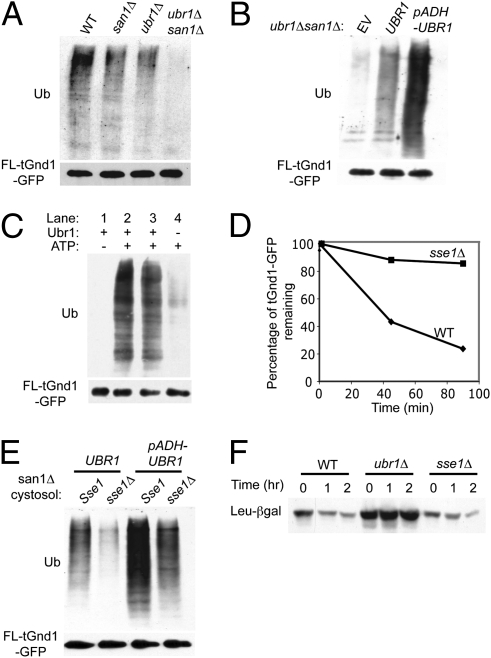
The observation that San1 can recognize a variety of misfolded substrates is consistent with its known role as a QC E3 (9). However, a role for Ubr1 in QC was unique and unexplored. Accordingly, we next examined mechanistic features of Ubr1-mediated QC in greater detail. We used the substrate tGnd1p-GFP to test more fully the idea that Ubr1-mediated QC was distinct from its well-studied role in the N-end rule, in which the N terminus of a protein determines its rate of Ubr1-mediated degradation (11, 23, 24). Ubr1 is the E3 ligase, or “N-recognin” of the N-end rule pathway, binding to and catalyzing ubiquitination of proteins with appropriate N-terminal amino acids present by cleavage or enzymatic addition (16). It was formally possible that the Ubr1-dependent ubiquitination of our misfolded substrates was attributable to cleavage of a small amount of the protein to reveal a fast-degradation N-terminal residue, followed by traditional N-end recognition by Ubr1. We addressed this issue in several ways. We expressed and immunoprecipitated FLAG-tGnd1-GFP, bearing a single N-terminal tag, with anti-FLAG antibody to evaluate the ability of Ubr1 to ubiquitinate tGnd1-GFP with an intact N terminus. The resulting ubiquitination in the suite of four strains was identical to that seen above with anti-GFP antibodies (Fig. 2D and Fig. 4A), with a similarly strong Ubr1-dependent component of ubiquitination, indicating that cleavage was not required for this ubiquitination. We developed an in vitro ubiquitination assay to test directly the ability of Ubr1 to ubiquitinate intact FLAG-tGnd1-GFP. The substrate was immunoprecipitated from a ubr1Δsan1Δ strain with anti-FLAG antibody beads. The beads were washed several times and then incubated with WT cytosol and added ATP. The beads were then washed, and the bound protein was evaluated for ubiquitination by immunoblotting as above (Fig. S3A). A strong ubiquitination signal was observed only when the beads with bound substrate were incubated in cytosol and added ATP (Fig. S3B). We used this assay to test directly Ubr1-mediated ubiquitination of full-length FLAG-tGnd1-GFP. Anti-FLAG bead-bound substrate, immunoprecipitated from a ubr1Δsan1Δ strain, was subjected to in vitro ubiquitination by cytosol from ubr1Δsan1Δ strains with no Ubr1 (empty vector), plasmid-expressing native promoter Ubr1 (UBR1), or overexpressed Ubr1 (pADH-UBR1). The FLAG-tGnd1-GFP bound by its intact N terminus was ubiquitinated in vitro in a Ubr1-dependent manner, and Ubr1 was rate-limiting (Fig. 4B). To ascertain whether Ubr1 was directly involved in tGnd1-GFP ubiquitination, we examined the effects of immunodepletion from the reaction cytosol using FLAG-Ubr1 as the E3. Anti-FLAG precipitation of Ubr1 from cytosol immediately before the ubiquitination assay resulted in complete inhibition of ubiquitination, whereas no effect was observed with nonspecific control beads (Fig. 4C). Taken together, these results demonstrated that N-terminal cleavage was not required for Ubr1-dependent ubiquitination of tGnd1-GFP and that Ubr1-mediated ubiquitination of QC substrates is directly mediated by this ligase.
Fig. 4.
Ubr1 acts independent of the N-end rule in cytoplasmic QC. (A) IP of FLAG-tGnd1-GFP with anti-FLAG antibody from the indicated strains, followed by immunoblotting with antiubiquitin (Ub) or anti-GFP. (B) Ubr1 dependence of in vitro tGnd1-GFP ubiquitination. Immunopurified FLAG-tGnd1-GFP was incubated with cytosol from ubr1Δsan1Δ strains expressing empty vector (EV), native promoter-driven UBR1, or pADH-UBR1 as indicated and was incubated for 1 h at 30°C and evaluated for tGnd1-GFP ubiquitination. (C) In vitro ubiquitination of bead-bound tGnd1-GFP after depletion of FLAG-UBR1 from san1Δ cytosol. Reaction cytosols were either untreated (lanes 1 and 2) or preincubated with IgG agarose beads as a control (lane 3) or with anti-FLAG beads (lane 4). “Depleted” cytosols were then added to substrate-bound beads, with or without ATP, and ubiquitination was assayed as above. (D) In vivo degradation of FLAG-tGnd1-GFP in WT or sse1Δ by flow cytometry, normalized to the mean fluorescence at time 0. (E) Sse1 requirement for Ubr1-dependent in vitro ubiquitination of tGnd1-GFP. Bead-bound tGnd1 was incubated in san1Δ null cytosols with or without Sse1. Left-pair strains expressed Ubr1 at native levels, and right-pair strains expressed Ubr1p from the strong ADH promoter. (F) Cycloheximide chase of N-end rule substrate Leu-β-gal in WT, ubr1Δ, or sse1Δ strains, immunoblotted with anti-β-gal antibodies.
The QC function of Ubr1 can be further distinguished from its role in the N-end rule through its clear chaperone dependence. A defining feature of Ubr1-mediated QC in our studies is dependence on molecular chaperones for both degradation and ubiquitination (see below). In particular, all our QC substrates reveal a strong dependence on the Sse1 chaperone (Fig. 4D and Fig. S2 B, C, and E). This was contrary to earlier published work concerning Sse1 and CPY‡-GFP (14). Using the in vitro assay, we directly tested the role of Sse1 in Ubr1-mediated ubiquitination of tGnd1. Bead-bound FLAG-tGnd1-GFP was incubated in san1Δ cytosol with WT or overexpressed Ubr1, with or without Sse1 (Fig. 4E). Ubr1-dependent ubiquitination of FLAG-Gnd1p-GFP was strongly decreased in the sse1Δ cytosol at both levels of Ubr1. Thus, in vivo and in vitro Ubr1 QC function is dependent on Sse1. This is another criterion that distinguishes Ubr1-mediated QC from the N-end rule: no chaperone dependence of the classic N-end rule pathway has been reported, implying that the chaperone dependence is a unique feature of Ubr1-mediated QC. We confirmed this by testing the effect of the sse1Δ null on the classic N-end rule substrate, Leu-β-gal (23). This substrate showed identical degradation in WT or sse1Δ strains, but the expected strong stabilization in an isogenic ubr1Δ strain was observed (Fig. 4F).
N-end rule-dependent degradation can also occur when a “fast” N terminus is added to a stable N terminus by the action Ate1 arginyl transferase (25). Accordingly, we tested Ubr1-dependent degradation in ate1Δ and ate1Δsan1Δ nulls (Fig. S4). No effect was observed in ate1Δ strains, indicating that the Ubr1-dependent degradation of these misfolded proteins did not require addition of arginine at the N terminus. Taken together, these experiments demonstrated that Ubr1-mediated recognition of QC substrates was independent of its well-described function in the N-end rule.
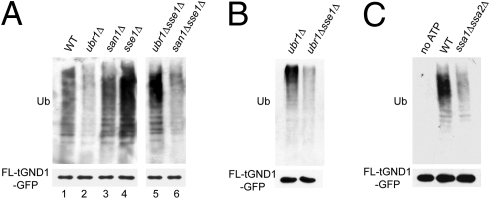
We used the in vitro assay to evaluate the role of chaperones in Ubr1- and San1-mediated QC. In vivo, loss of either Sse1 or Hsp70 caused a strong defect in ubiquitination, implying that they were involved in the action of each ligase. However, our in vitro analysis indicated that there are clear differences in the role of chaperones with each ligase. In vivo, FLAG-tGnd1-GFP ubiquitination was mediated by either Ubr1 or San1 (Fig. 4A). However, in vitro, the same substrate was ubiquitinated almost entirely by Ubr1. A ubr1Δ null cytosol was incapable of supporting tGnd1-GFP ubiquitination, whereas the reaction in a san1Δ cytosol was identical to that in WT (Fig. 5A, lanes 1–3). What is the cause of this difference in vitro? We discovered that the strong in vitro bias toward Ubr1-dependent tGnd1 ubiquitination was attributable to a surprising inhibition of San1 by Sse1. An sse1Δ cytosol supported robust ubiquitination of the substrate (Fig. 5A, lane 4). This sse1Δ-stimulated ubiquitination was strongly dependent on San1 (Fig. 5A, lane 6), showing a large decrease in the sse1Δsan1Δ double null. This is in striking contrast to the case in the intact cell, in which the San1-dependent component of tGnd1-GFP ubiquitination (measured in a ubr1Δ null) was strongly inhibited by the presence of the sse1Δ null (Fig. 5B).
Fig. 5.
Differential chaperone requirement for Ubr1- and San1-mediated ubiquitination. (A) In vitro ubiquitination of tGnd1-GFP bound to anti-FLAG beads after incubation with cytosol from WT, ubr1Δ, san1Δ, sse1Δ, ubr1Δsse1Δ, or san1Δsse1Δ strains. Ub, ubiquitin. (B) In vivo ubiquitination of FLAG-tGnd1-GFP in ubr1Δ or ubr1Δsse1Δ strains. FLAG-tGnd1-GFP was immunoprecipitated from cell lysates with anti-FLAG beads and immunoblotted for GFP or Ub. (C) In vitro ubiquitination of FLAG-tGnd1-GFP. tGnd1-GFP bound to anti-FLAG beads was incubated with cytosol from WT or ssa1Δssa2Δ strains for 1 h at 30°C and assayed for ubiquitination by immunoblotting for GFP or Ub.
Our in vivo studies indicate that Ubr1-mediated QC is Hsp70-dependent. The in vitro assays above (Fig. 4E) show that Ubr1-mediated ubiquitination depends on the Sse1 cochaperone. Because the in vitro assay is almost entirely Ubr1-dependent in these conditions, we used this assay to examine the role of Hsp70 in Ubr1 QC directly. Indeed, loss of two of the four redundant Hsp70 genes (ssa1Δssa2Δ) resulted in a strong decrease in Ubr1-mediated tGnd1-GFP ubiquitination in vitro (Fig. 5C), indicating a direct role for Hsp70 in this unique function of Ubr1. Taken together, these studies indicate that Ubr1 directly employs chaperones for ubiquitination of substrates, whereas San1 has a more complex relation with chaperones that appears to operate separately from substrate ubiquitination (see below).
These studies have defined Ubr1 and San1 as E3 ligases involved in chaperone-dependent cytoplasmic QC. Degradation mediated by these E3s meets the criteria for physiologically relevant protein QC: ubiquitination is selective for misfolded proteins, a broad range of misfolded substrates are recognized by either ligase, and both ligases participate in management of proteotoxic stress. Each ligase has been studied in other capacities. Ubr1 is known for degradation of N-end rule substrates as well as numerous proteins recognized by distinct mechanisms (11, 12). San1 is a recently described nuclear QC E3 (9). Our work thus significantly extends the functional range of each ligase.
The previously unknown QC function for Ubr1 was distinct from its elegantly described action in the N-end rule and was restricted to the Ubr1 isozyme. Both E2s reported to work with Ubr1 could support ubiquitination of a Ubr1-selective QC substrate. Intriguingly, Ubr1 has a number of reported substrates that are not recognized by the N-end rule, including some that bear degradation determinants also found in misfolded proteins (26–28). It will be important to reevaluate these proteins as possible QC substrates. In addition, a variety of phenotypes have been observed in murine nulls of several Ubr isoforms (29–32); it may be that these effects are the result of deficient QC control in the affected tissues.
Although the San1 ubiquitin ligase was originally reported as a QC E3 (9), our studies significantly expand the range of substrates to include misfolded proteins of cytoplasmic origin. Because San1 originates in the cytosol, it was possible that the cytoplasmic QC function was attributable to the subset of molecules present in that compartment at steady state. Indeed, San1 restricted to the cytosol was fully capable of mediating degradation of CPY‡-GFP. However, the other studies in this work imply that San1 and Ubr1 function in distinct compartments to mediate destruction of misfolded cytoplasmic proteins (see below).
It has been previously reported that degradation of cytoplasmic proteins requires chaperones (5, 14), and the Ubr1/San1-dependent substrates appropriately showed a strong dependence on Hsp70 and the Sse1 (Hsp110) cochaperone. However, the role of chaperones in the action of each was distinguishable. Ubr1 appeared to use both Sse1 and Hsp70 directly, as indicated by the in vitro assays. In contrast, although San1-dependent degradation (observed in a ubr1Δ null) was strongly Sse1-dependent in vivo, its action was inhibited by Sse1 in vitro.
Taken together, our data suggest a two-compartment model for the roles of Ubr1 and San1 in cytoplasmic QC. The degradation and phenotypic studies with the cytoplasm-restricted CPY‡-GFP+NES were consistent with distinct actions of these ligases. Ubr1 maintained its ability to degrade this purely cytoplasmic substrate, whereas San1 had no role in its degradation. Similarly, the stress phenotype caused by this cytoplasmic substrate was only sensitive to the ubr1Δ and not to the san1Δ, again indicating a direct cytoplasmic role for Ubr1 but not for San1. Taken together, these data suggest the following model. A misfolded cytoplasmic protein can undergo ubiquitination by either Ubr1 or San1. The Ubr1-dependent branch occurs in the cytosol, using chaperones in conjunction with Ubr1 to recognize and ubiquitinate a substrate. In contrast, San1-dependent ubiquitination of the same substrate requires chaperone-dependent transport into the nucleus, where San1 ubiquitinates the substrate. The dichotomous requirements for Sse1 observed for San1 in vivo and in vitro are consistent with this model: in vivo, Sse1, in conjunction with Hsp70, promotes delivery of misfolded substrate to the nuclear pool of San1. In vitro, Sse1 gains access to San1 and inhibits it, perhaps by interacting with San1 features that interact with misfolded proteins. In support of this idea, we have observed that a fraction of CPY‡-GFP builds up in the nucleus in a san1Δ null and that this buildup is dependent on Sse1 (Fig. S5): In an sse1Δ san1Δ double null, CPY‡-GFP accumulates in the cytoplasm only. This dependency appeared to be strong, because addition of the NES construct to the CPY‡-GFP had almost the same effect as sse1Δ in this optical experiment (Fig. S5). Direct tests of this model will be an important avenue of future inquiry.
Our studies with nulls indicate that the two pathways function independently: Each E3 appears to contribute a component of ubiquitination that is independent of the presence of the other ligase. However, there was phenotypic synergy in the stress experiments involving CPY‡-GFP, because loss of both E3s had a much more drastic effect than that of either single null (Fig. 3A). Thus, the issue of regulatory or mechanistic crosstalk between these two modes of cytoplasmic QC is an open and interesting one. It is clear that Ubr1 plays the principal role in both degradation of cytoplasmic substrates and management of cytoplasmic stress. Another open question is the role of chaperones in the QC action of Ubr1. This includes discerning the full panoply of chaperones and cochaperones involved and understanding the mechanism by which they assist Ubr1 in substrate ubiquitination. It could be that they are required to produce substrate forms that are recognizable by Ubr1. Alternatively, it may be that Hsp70 and Sse1 form a substrate recognition module that works directly with Ubr1, in a manner analogous to CHIP (7). Finally, we have observed that some substrates that undergo chaperone-dependent degradation are not subject to Ubr1/San1 ubiquitination (5), indicating that other E3 QC ligases remain to be discovered. In addition, it is clear that the Doa10 ERAD ligase can participate in recognition of some cytoplasmic QC substrates (2). Thus, the full picture of cytoplasmic QC will involve a network of interacting options.
A large number of clinically pressing maladies have etiologies that pertain to protein misfolding, including aging, parkinsonism, Huntington’s disease, and a variety of somatic illnesses (33). Identification of the relevant QC ligases opens numerous doors to understanding pathological characteristics of proteostasis. Detailed knowledge of these QC pathways will allow their manipulation for basic and clinical purposes.
Materials and Methods
Yeast Strains and Plasmids.
The Saccharomyces cerevisiae strains used in this work are listed in Table S1. All strains and plasmids were constructed with standard molecular biology techniques, as described elsewhere (34, 35).
In Vitro Ubiquitination Assay.
In vitro ubiquitination assays were adapted from Garza et al. (35). Briefly, bead-bound substrate was mixed with cytoplasm isolated from the indicated genetic background. One hundred fifty milligrams of total protein was mixed with 15 mM ATP and 10 μL of substrate-bound beads. The reaction was incubated at 30°C for 1 h. Bead-bound substrate was then washed several times with immunoprecipitation (IP) buffer, protease inhibitors, and N-ethylmaleimide; it was then aspirated to dryness and subjected to electrophoretic sample buffer and immunoblotting. A more detailed description is provided in SI Text.
In Vivo Ubiquitination Assay.
In vivo ubiquitination of substrates was evaluated by IP, followed by ubiquitin immunoblotting as described by Bays et al. (37). Cells were lysed in the presence of protease inhibitors and N-ethylmaleimide, followed by IP of the target substrate with anti-GFP, anti-HA (Covance), or anti-FLAG M2 beads (Sigma-Aldrich). After several washes, electrophoretic sample buffer was added, followed by immunoblot analysis. A more detailed description is provided in SI Text.
Supplementary Material
Acknowledgments
We thank David Ng and Richard Gardner for sharing unpublished data relevant to this study. We also thank Dieter Wolf (University of Stuttgart, Stuttgart, Germany), Alex Varshavsky (California Institute of Technology), Richard Gardner (University of Washington), and Davis Ng (Tamasek Life Sciences Institute, Singapore) for strains and/or plasmids. As always, we thank the Hampton Laboratory for enthusiastic interactions of an experimental, psychological, recreational, and theoretical nature. This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK051996 (to R.Y.H.) and by an American Heart Association Established Investigator Award (to R.Y.H.). J.W.H. was supported in part by the University of California, San Diego, Genetics Training Program (Grant T32 GM008666 from the National Institute of General Medical Sciences).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910591107/DCSupplemental.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vashist S, et al. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhovsky S, Gardner R, Hampton R. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 7.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta A, Ramsey KL, Smith JS, Auble DT. Sir Antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem. 2004;279:26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- 11.Varshavsky A. Recent studies of the ubiquitin system and the N-end rule pathway. Harvey Lect. 2000-2001;96:93–116. [PubMed] [Google Scholar]
- 12.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SH, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 16.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hampton RY. Fusion-based strategies to identify genes involved in degradation of a specific substrate. Methods Enzymol. 2005;399:310–323. doi: 10.1016/S0076-6879(05)99021-6. [DOI] [PubMed] [Google Scholar]
- 18.Turnbull EL, Rosser MF, Cyr DM. The role of the UPS in cystic fibrosis. BMC Biochem. 2007;8(Suppl 1):S11. doi: 10.1186/1471-2091-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He W, Wang Y, Liu W, Zhou CZ. Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1. BMC Struct Biol. 2007;7:38. doi: 10.1186/1472-6807-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, et al. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 23.Varshavsky A, Bachmair A, Finley D. The N-end rule of selective protein turnover: Mechanistic aspects and functional implications. Biochem Soc Trans. 1987;15:815–816. doi: 10.1042/bst0150815. [DOI] [PubMed] [Google Scholar]
- 24.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 25.Hu RG, et al. Arginyltransferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J Biol Chem. 2006;281:32559–32573. doi: 10.1074/jbc.M604355200. [DOI] [PubMed] [Google Scholar]
- 26.Hwang CS, Shemorry A, Varshavsky A. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc Natl Acad Sci USA. 2009;106:2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madura K, Varshavsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 28.Lawson TG, Sweep ME, Schlax PE, Bohnsack RN, Haas AL. Kinetic analysis of the conjugation of ubiquitin to picornavirus 3C proteases catalyzed by the mammalian ubiquitin-protein ligase E3alpha. J Biol Chem. 2001;276:39629–39637. doi: 10.1074/jbc.M102659200. [DOI] [PubMed] [Google Scholar]
- 29.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenker M, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. and correction (2006) 38:265. [DOI] [PubMed] [Google Scholar]
- 31.Tasaki T, et al. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- 32.An JY, et al. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powers ET, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 34.Gardner R, et al. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1998;9:2611–2626. doi: 10.1091/mbc.9.9.2611. and correction (1999) 10:precede. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardner RG, Hampton RY. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.