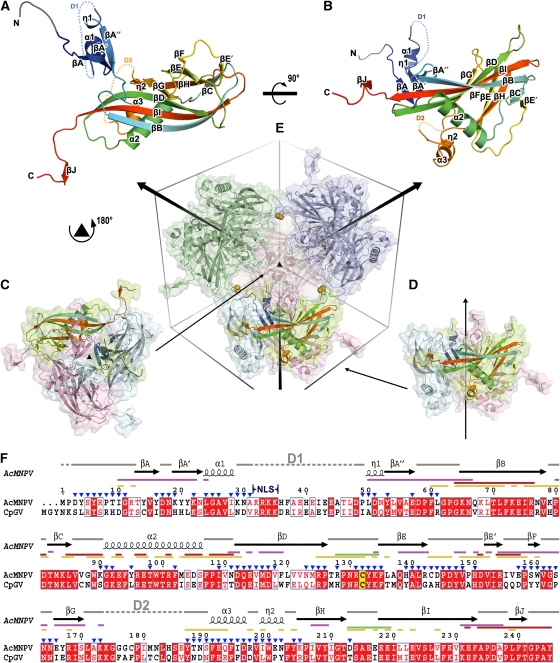
Figure 1.
Overview of structure. (A, B) The structure of AcMNPV polyhedrin, in two orthogonal views, shown as cartoon representations coloured from blue at the N-terminus to red at the C-terminus. Residues 3–7, which were built as poly-ala are drawn in grey. Disordered segments D1 (residues 32–48) and D2 (residues 174–186) are shown as dots. (C, D) Cartoon and surface representations of polyhedrin trimers shown in two orientations aligned to (E). Subunits are coloured by chain with one in each trimer coloured as in (A). (E) Dodecameric unit of four trimers linked by disulphide bonds (represented as orange spheres). The unit cell and three-fold axes are drawn to facilitate observation. (F) Amino-acid sequence conservation between the type species of the alphaBVs (AcMNPV) and betaBVs (CpGV). Conserved residues drawn in red boxes, similar residues in red type. The secondary structure assignment for AcMNPV is drawn above. Conserved cysteine is highlighted in yellow. Interactions involved in various stages of oligomization are mapped as lines coloured thus; purple for trimer, green for dodecamer, red for interactions through C-terminal and yellow for other interactions in the unit cell. The position of the nuclear localization signal (NLS) is indicated and residues exposed to the cavity are marked by blue triangles. The sequences were aligned with ClustalW (Chenna et al, 2003) and visualized using ESPript (Gouet et al, 1999).