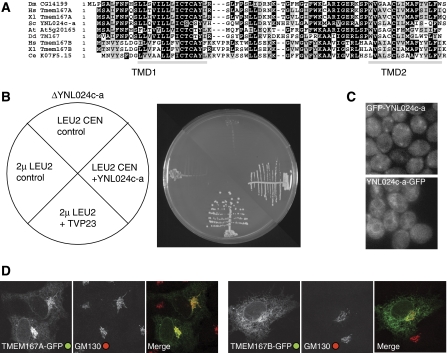
Figure 7.
Analysis of the yeast and mammalian orthologues of CG14199/Kish. (A) An alignment of CG14199/Kish with orthologues from the indicated species (Hs, Homo sapiens; Xl, Xenopus laevis; Sc, S. cerevisiae; At, Arabidopsis thaliana; Dd, Dictyostelium discoideum; Ce, Caenorhabditis elegans). Shading indicates residues that are identical (black) or conserved (grey) in five or more of the nine sequences, and with grey bars being predicted transmembrane domains. Vertebrates have two paralogues of Kish called TMEM167A and TMEM167B. (B) Growth of S. cerevisiae with a deletion in YNL024c-a (KSH1), the orthologue of CG14199. Strains were created that lacked the genomic copy of YNL024c-a but were kept alive by the YNL024c-a gene on a URA3 CEN ‘covering' plasmid, and also contained the indicated CEN (pRS415) or 2 μ LEU2 plasmids. The strains were streaked on 5-fluororotic acid (5-FOA) to eject the URA3 covering plasmid and then incubated for 3 days at 25°C before photography. The LEU2 plasmids containing YNL024c-a and TVP23 expressed from their own promoters support growth in the absence of genomic YNL024c-a, but this was not seen with the control plasmids (an empty CEN plasmid, or a 2μ plasmid expressing an irrelevant gene (LRC6)). (C) Wide-field micrographs of wild-type strain BY4741 with GFP integrated at the N or C terminus of the endogenous YNL024c-a gene by PCR-based homologous recombination. For the N-terminal fusion expression was promoted by a constitutive form of the PHO5 promoter. (D) Confocal micrographs of COS cells expressing the human orthologues of Kish with GFP attached to the C terminus, and labelled for the Golgi marker GM130.