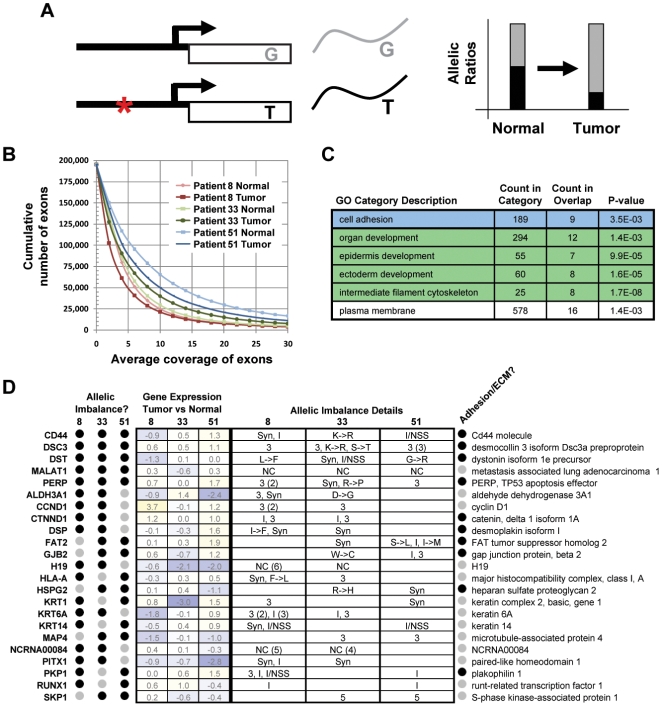
Figure 4. A common set of genes, functioning in cell adhesion and development, exhibits allelic imbalance in the tumor transcriptomes of three patients with oral carcinoma.
Allelic imbalance (AI) at the RNA level can arise in a number of ways, including through point mutation and changes in the relative expression of alleles (aka, allele-specific expression). (A) Illustrated is an example of two pre-existing alleles (G and T), one of which undergoes a linked promoter mutation (red asterisk) in the tumor, relative to the normal tissue. If, for example, the mutation alters a cis-regulatory element, then the balance of the two alelles (G and T) may change. (B) In principle, there is enough sequence coverage to discover AI for a large number of exons. For example, more than 25,000 exons have at least 10x coverage (averaged across all sites in the exon). (C) Genes with one or more instances of allelic imbalance in the tumors of two or more patients were submitted for GO analysis [29] to identify biological processes and components that are typically allelically imbalanced in oral cancer. Redundant GO categories were filtered. (D) A selection of genes with allelic imbalance in two or more patients is listed, along with the log2 differential expression of each gene and regions of gene structure impacted. Also noted is whether or not the gene is involved in cell adhesion or is a component of the ECM. Key: 3 = 3′ UTR, I = Intronic, NSS = Near Splice Site, Syn = Synonymous, → = Non-Synonymous.