Figure 3.
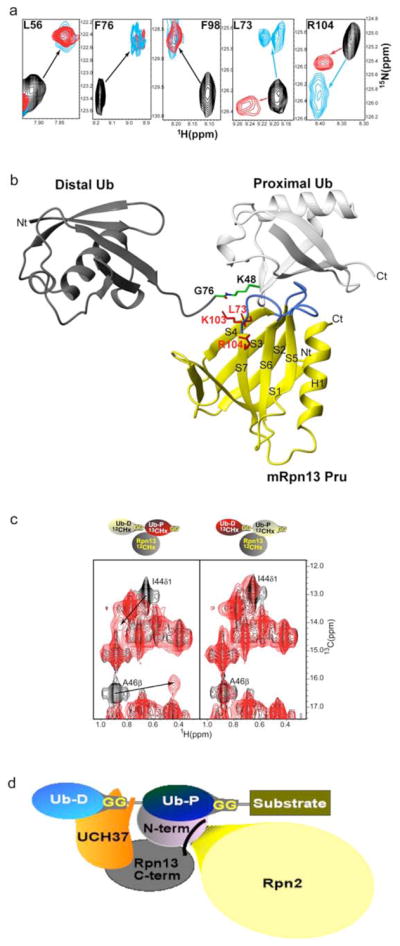
a, Comparison of spectra acquired with monoubiquitin versus K48-linked tetraubiquitin reveal identical affects for Rpn13’s L56, F76 and F98 but differences for L73 and R104. Monoubiquitin (red) or tetraubiquitin (cyan) was added to hRpn13, which was monitored by 1H, 15N HSQC experiments. The spectrum of free hRpn13 is indicated in black, and the molar ratio of monoubiquitin (red) or tetraubiquitin (cyan) to hRpn13 was 1:1 in the represented spectra.
b, Computer-generated model of the mRpn13 Pru:diubiquitin complex. White and grey ribbon diagrams display the proximal and distal ubiquitin, respectively, whereas a balls-and-sticks representation is used for the K48–G76 isopeptide bond linkage. Rpn13 Pru is colored in yellow and loops recognizing ubiquitin in blue, whereas L73, K103 and R104 are displayed in red. Diubiquitin was created by Insight II software based on atomic coordinates for the mRpn13 Pru:ubiquitin complex and monoubiquitin (PDB entry 1D3Z). In this model, the distal subunit of diubiquitin is positioned arbitrary, as its only constraints prohibit steric clashes with other atoms.
c, hRpn13 Pru interacts with the I44 δ1 and A46 methyl groups of the proximal subunit. 1H, 13C HMQC spectra were acquired on samples containing either no (black) or equimolar unlabeled hRpn13 (1–150) (red) mixed with diubiquitin in which its proximal (left) or distal (right) subunit is 13C labeled. The shifted resonances are labeled.
d, Model for how Rpn13 participates in Uch37 deconjugation of ubiquitinated substrates. Rpn13’s C-terminal domain (grey) binds Uch37 (orange) as its N-terminal domain binds the polyubiquitin chain and Rpn2/S1 (yellow). In this model, Uch37 binds to the distal subunit (light blue) of the chain while Rpn13 binds the proximal subunit (dark blue).
