Abstract
Background
Gene expression analysis has many applications in cancer diagnosis, prognosis and therapeutic care. Relative quantification is the most widely adopted approach whereby quantification of gene expression is normalised relative to an endogenously expressed control (EC) gene. Central to the reliable determination of gene expression is the choice of control gene. The purpose of this study was to evaluate a panel of candidate EC genes from which to identify the most stably expressed gene(s) to normalise RQ-PCR data derived from primary colorectal cancer tissue.
Results
The expression of thirteen candidate EC genes: B2M, HPRT, GAPDH, ACTB, PPIA, HCRT, SLC25A23, DTX3, APOC4, RTDR1, KRTAP12-3, CHRNB4 and MRPL19 were analysed in a cohort of 64 colorectal tumours and tumour associated normal specimens. CXCL12, FABP1, MUC2 and PDCD4 genes were chosen as target genes against which a comparison of the effect of each EC gene on gene expression could be determined. Data analysis using descriptive statistics, geNorm, NormFinder and qBasePlus indicated significant difference in variances between candidate EC genes. We determined that two genes were required for optimal normalisation and identified B2M and PPIA as the most stably expressed and reliable EC genes.
Conclusion
This study identified that the combination of two EC genes (B2M and PPIA) more accurately normalised RQ-PCR data in colorectal tissue. Although these control genes might not be optimal for use in other cancer studies, the approach described herein could serve as a template for the identification of valid ECs in other cancer types.
Background
Colorectal cancer (CRC) is one of the most common causes of cancer worldwide affecting almost a million people annually and resulting in approximately 500,000 deaths [1]. Approximately 5% of individuals born today will be diagnosed with colorectal cancer during their lives, representing a lifetime risk of 1 in 19. CRC remains a serious threat to life with approximately 20% of patients presenting with late stage metastatic disease. Although 5 year survival rates are favourable at 80-90% for early stage disease, this drops significantly to less than 10% with the presence of distal metastasis.
The majority of colorectal tumours originate from adenomatous precursor lesions and develop along a well-defined adenoma-carcinoma sequence. According to this model the culmination of mutational events including activation of oncogenes and loss of function of tumour suppressor genes results in the emergence of carcinomas [2]. Molecular profiling across the spectrum of normal-adenoma-tumour tissue types has yielded many candidate genes in the search for novel molecular diagnostic and prognostic markers and treatment strategies [3-5]. In latter years real-time quantitative (RQ-) PCR has become established as the gold standard for accurate, sensitive and rapid quantification of gene expression [6,7]. In comparison to alternative methods such as Northern blotting and Ribonuclease Protection Assays (RPA), RQ-PCR has been universally adopted as the transcriptomic method of choice due to its superiority with regard to speed, sensitivity, reproducibility and the wide range of instrumentation and reagents commercially available.
To accurately quantify an mRNA target by RQ-PCR, samples are assayed during the exponential phase of the PCR reaction during which the amount of target is assumed to double with each cycle of PCR without bias due to limiting reagents. Analysis of cycle threshold (Ct), the cycle number at which signals are detected above background, can be used to estimate gene expression levels by relating Ct values either to a standard curve (absolute quantification) or to a control gene (relative quantification). The latter method requires the generation of standard curves of known copy number for each target and so is limited due to logistical issues associated with the generation of standards in studies of multiple gene targets. Relative quantification is the most widely adopted approach and as the name suggests, quantification of gene expression is based on the analysis of a target gene whose expression is normalised relative to the expression of a control gene. Central to the reliable determination of gene expression is the choice of control gene with which to normalise real-time data from target genes. Normalisation can be achieved using endogenous or exogenous controls; however the use of endogenous control (EC) genes is the most widely adopted approach as it excludes variation associated with differences in amounts of template RNA. Vandestompele et al 2002 described a normalisation method whereby geometrical averaging of multiple EC genes improved accuracy [8]. This approach has been adopted to reliably measure levels of gene expression in many studies in different tissue types including breast [9-11], lung [12], kidney [13], brain [14] and liver [15].
An ideal EC gene (or genes) should be stably expressed and unaffected by parameters such as disease status and in the case of CRC, should remain unaffected by whether a tissue was derived from normal, adenoma or carcinoma lesions. Traditionally GAPDH (glyceraldehyde phosphate dehydrogenase) has been widely used to normalise RQ-PCR data. A common feature of earlier studies was that the stability of reference gene expression between different sample types was assumed with little consideration paid to validation of these EC genes as suitable normalisers. More recent studies have brought into question the stability of commonly used EC genes such as GAPDH on the basis that gene expression levels have been found to vary in response to treatment or as a result of physiological, pathological or experimental changes. For example, alteration in oxygen tension and hypoxia were found to be associated with wide variation in GAPDH, B-ACTIN and CYCLOPHILIN expression [16]. In addition, GAPDH expression was found to be strongly unregulated in diabetic patients and down-regulated in response to the administration of bisphosphonate compounds in the treatment of metastatic breast cancer [17]. Other evidence indicates that neoplastic growth can affect EC expression levels [18]. Goidin et al [19] found differences in the expression of GAPDH and B-ACTIN in two sub-populations of melanoma cells derived from a tumour in a single patient. Treatment agents such as dexamethasone, deprenyl and isatin also affect EC gene expression [20,21]. Schmittgen et al [22] reported increased expression of GAPDH, B2M, 18S rRNA and β-ACTIN in fibroblasts after the addition of serum: evidence of the effect of experimental conditions on EC expression. These findings were further supported by Wu et al [23] in their investigation of the effect of different skin irritants on GAPDH and PolyA+ RNA expression. GAPDH was found to be involved in age-induced apoptosis in mature cerebellar cells [24] and also as a tRNA binding protein present in the nuclei of HeLa cells [25].
As the use of unreliable ECs can result in inaccurate results, the identification of the most reliable gene or set of genes at the outset of an investigation is critical. Thus far, a pervasive stably expressed gene (or genes) has yet to be identified across all tissue types [26,27]. This would indicate that the identification of robust ECs at the outset of transcriptomic analysis would yield more reliable and meaningful RQ-PCR data.
The aim of this study was to evaluate a panel of thirteen candidate EC genes from which to identify the most stably expressed gene (or genes) to normalise RQ-PCR data derived from primary colorectal tumour and tumour associated normal (TAN) tissue. Six of the candidate EC genes were selected from the literature and represent the most frequently studied reference genes in cancer including, but not limited to, colorectal cancer. Each gene was previously reported as being constitutively expressed in various tissues. These EC genes included B2M (beta-2-microglogulin) [5], HPRT (hypoxanthine guanine phosphoribosyl transferase 1) [3,28], GAPDH [29], ACTB (beta-actin) [30], PPIA (peptidyl-prolyl isomerise A) [9] and MRPL19 (mitochondrial ribosomal protein L19) [9]. The remaining seven genes included HCRT, SLC25A23, DTX3, APOC4, RTDR1, KRTAP12-3, and CHRNB4. The latter candidates were selected from an unpublished whole genome microarray dataset of 20 human tumour specimens and represented the most stably expressed probes with a fold-change of 1.0-1.2, (p < 0.05). Expression of CXCL12 [31], FABP1 [32], MUC2 [33] and PDCD4 genes were chosen as targets against which to measure the effects of candidate EC expression on the basis of their previously identified roles in tumourigenesis. In addition to its tumour suppressor properties, PDCD4 [34] also has diagnostic and prognostic utility and represents a promising target for anti-cancer therapy.
Results
Range of Expression of Candidate EC Genes
A range of Ct values was observed across the candidate EC genes in tumour and TAN tissue from CRC patients as indicated in table 1. Only samples with a standard deviation < 0.3 from the mean Ct of the triplicates were included for further analysis. The expression of RTDR1, HCRT, APOC4 and KRTAP12-3 could not be determined in all 64 tissue samples, resultantly these candidates were excluded from further analysis.
Table 1.
Clinico-pathological patient data for tumour and tumour associated normal colorectal tissues
| Clinicopathological Variable | Number of Patients N = 42 |
|---|---|
| Gender | |
| Males | 29 |
| Females | 13 |
| Mean Age (SD) | 66.5 (12.84) |
| Tumour Location | |
| Colon | 12 |
| Rectum | 30 |
| Tumour Diameter (mm) | |
| <10 | 11 |
| 10-15 | 15 |
| >15 | 17 |
| Tumour Thickness (mm) | |
| <30 | 15 |
| 30-40 | 12 |
| >40 | 15 |
| Distant Metastasis | |
| M0 | 36 |
| M1 | 6 |
| Nodal Status | |
| N0 | 22 |
| N1 | 11 |
| N2 | 9 |
| UICC Stage | |
| Stage 0 | 6 |
| Stage I | 10 |
| Stage II | 10 |
| Stage III | 11 |
| Stage IV | 5 |
| Tumour Differentiation | |
| Well | 12 |
| Moderate | 24 |
| Poor | 6 |
| Mucin Secretion | |
| Mucinous | 8 |
| Non-mucinous | 34 |
Mean Ct values for the remaining genes ranged from 19.48 (± 0.14 s.e.m) for B2M to 32.30 (± 0.19 s.e.m) for CHRNB4. B2M displayed the narrowest range of Ct values between 17.5 and 21.5 (mean 19.5 ± 0.14 s.e.m, range of 4.04) followed by PPIA and MRPL19, while ACTB had the widest range of Ct values between 33.8 and 21.1. The genes broadly fell into three categories, those least abundant genes with mean Ct values of 27-32 (SLC25A23, MRPL19, DTX3 and CHRNB4), moderately abundant genes with mean Ct values of 22-26 (HPRT and ACTB) and the most abundant highly expressed genes with mean Ct values of 19-21 (B2M, PPIA and GAPDH) Table 2.
Table 2.
Cycle threshold (Ct) values of candidate EC genes and PCDC4 in colorectal tissues
| EC Gene | Ct Range | Ct Min | Ct Max | Mean Ct ± s.e.m | Standard deviation (SD) |
|---|---|---|---|---|---|
| B2M | 4.03 | 17.47 | 21.51 | 19.48 ± 0.14 | 1.04 |
| PPIA | 4.13 | 17.78 | 21.91 | 19.90 ± 0.14 | 1.06 |
| GAPDH | 5.80 | 18.51 | 24.32 | 21.00 ± 0.17 | 1.29 |
| ACTB | 12.74 | 21.08 | 33.32 | 25.14 ± 0.34 | 2.61 |
| HPRT | 8.54 | 22.74 | 31.28 | 26.68 ± 0.25 | 1.89 |
| DTX3 | 6.6 | 24.95 | 31.56 | 28.62 ±0.17 | 1.37 |
| SLC25A23 | 7.26 | 24.48 | 31.74 | 27.36 ±0.19 | 1.54 |
| CHRNB4 | 9.40 | 27.99 | 37.38 | 32.30 ±0.19 | 2.15 |
| RTDR1 | - | 30.59 | UD | 35.82 ±0.36 | 2.15 |
| HCRT | - | 33.96 | UD | 38.46 ±0.29 | 1.67 |
| APOC4 | - | UD | UD | - | - |
| KRTAP12-3 | - | 33.16 | UD | 36.95 ±0.19 | 1.46 |
| MRPL19 | 4.10 | 26.70 | 30.80 | 28.62 ± 0.13 | 0.98 |
| CXCL12 | 13.54 | 21.85 | 35.39 | 25.77 ± 0.32 | 2.61 |
| FABP1 | 15.57 | 16.61 | 32.19 | 20.83 ± 0.40 | 3.24 |
| MUC2 | 17.71 | 17.62 | 35.33 | 22.43 ± 0.53 | 4.16 |
| PDCD4 | 11.92 | 21.35 | 33.27 | 24.56 ± 0.32 | 2.59 |
B2M and PPIA were the most abundantly expressed genes, having the lowest mean Ct values while MRPL19 was the least abundantly expressed gene with average Ct values > 26. Both B2M and PPIA had the lowest range in their Ct values.
Identification of Optimal EC genes
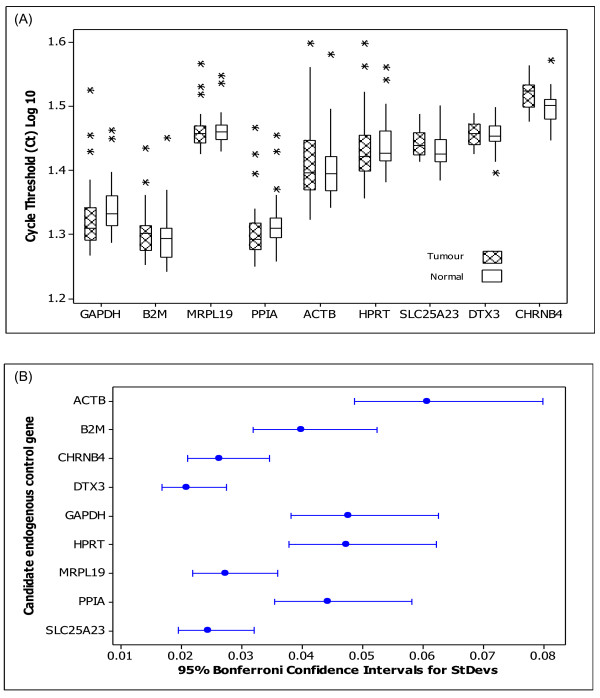
Scaled expression levels across the remaining nine candidate ECs analysed (figure 1) indicated within-gene differences in expression between tumour and normal tissue groups in both SLC25A23 (p = 0.040) and CHRNB4 (P = 0.002) but not in the remaining genes (p > 0.05), (figure 2A). Therefore, SLC25A23 and CHRNB4 genes were excluded from further analysis. Significant differences in variance of EC expression were identified using Levene's test (p < 0.001, figure 2B). These findings necessitated further evaluation of each candidate EC gene prior to their possible use to accurately quantitate gene expression levels of the target genes CXCL12, FABP1, MUC2 and PDCD4.
Figure 1.
Scaled expression levels and variation of each candidate EC gene. (A) Log 10 of cycle threshold of candidate EC genes ACTB, B2M, GAPDH, HPRT, MRPL19, SLC25A23, DTX3, CHRNB4 and PPIA in tumour and normal colorectal tissues. Boxplot shows interquartile range box, median, range whiskers and outliers (*). Within gene, differences were found in expression between tissue groups in both SLC25A23 (p = 0.040) and CHRNB4 (p = 0.002) but not the other genes (p > 0.05) (ANOVA). (B) Variation associated with EC gene expression. There was a significant difference in variation associated with gene expression (p < 0.001) with ACTB, GAPDH and HPRT showing greater variation than B2M, MRPL19 or PPIA. DTX3, CHRNB4 and SLC25A23 showed the least variations (Levene's test).
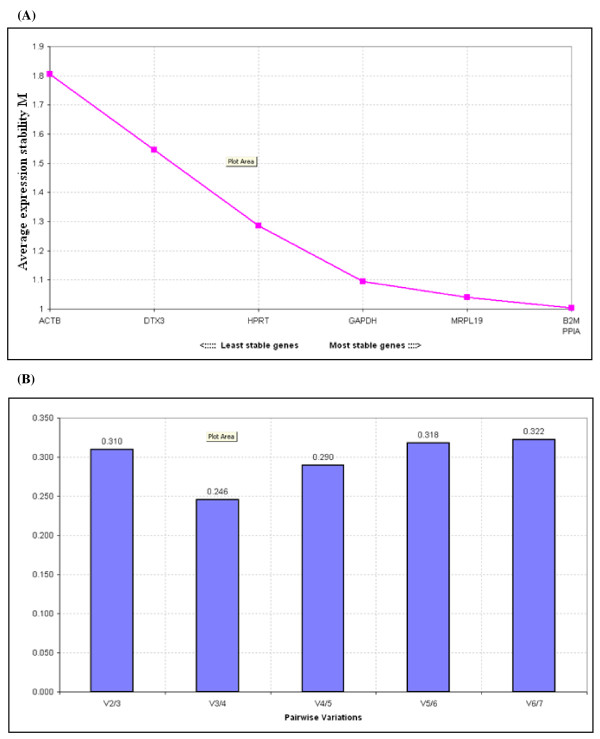
Figure 2.
Analysis of candidate EC genes using geNorm. (A): Average expression stability values of eligible EC genes. Expression stability of the control genes as calculated by geNorm. Stability value M is based on the average pair-wise variation between all genes. The least stable gene with highest M value was excluded and M value recalculated till end up with the most stable pair. (B): Determination of optimal number of control genes for normalisation. The GeNorm programme calculates a normalisation factor (NF) which is used to determine the optimal number of EC genes required for accurate normalisation. This factor is calculated using the variable V as the pairwise variation (Vn/Vn + 1) between two sequential NFs (NFn and NFn + 1). To meet the recommended cut off V-value which is the point at which it is unnecessary to include additional genes in a normalisation strategy. The recommended limit for V value is 0.15 but it is not always achievable. In this instance, the GeNorm output file indicated that the optimal number of genes required for normalisation was three.
The stability of candidate EC genes was analysed using geNorm [8] and NormFinder [35] programmes. Stability was further evaluated using qBasePlus [8,36], a commercially available RQ-PCR data mining package. These programmes were used to calculate amplification efficiency-corrected relative quantities from raw fluorescence data. The ranking of candidate EC genes as determined by each of these programmes is illustrated in Table 3. In the case of GeNorm the variable V indicating the pairwise variation (Vn/Vn+1) between two sequential normalisation factors (NFn/NFn+1) indicated that three EC genes was the optimal number of genes for accurate normalisation (figure 2), however, target genes expression did not differ significantly if two rather than three EC genes were used (figure 3). Use of all three programmes confirmed that B2M and PPIA was the best combination of genes for normalising RQ-PCR data in CRC tissues (table 3). The Equivalence test [37] was used to examine the expression of candidate ECs. All genes were equivalently expressed between the normal and tumour colorectal tissues using a fold cut-off of 2 (figure 4).
Table 3.
Ranking and best combination of EC genes as determined by geNorm, NormFinder and qBasePlus.
| Rank | GeNorm | NormFinder | qBaseplus | |||
|---|---|---|---|---|---|---|
| Gene | Stability (M) | Gene | Stability (M) | Gene | CV value | |
| 1 | GAPDH | 1.477 | MRPL19 | 0.008 | GAPDH | 0.555 |
| 2 | MRPL19 | 1.467 | B2M | 0.015 | PPIA | 0.659 |
| 3 | PPIA | 1.535 | HPRT | 0.016 | HPRT | 0.775 |
| 4 | B2M | 1.636 | PPIA | 0.017 | MRPL19 | 0.914 |
| 5 | HPRT | 1.813 | GAPDH | 0.018 | B2M | 0.923 |
| 6 | DTX3 | 2.251 | DTX3 | 0.020 | ACTB | 0.957 |
| 7 | ACTB | 2.454 | ACTB | 0.026 | DTX3 | 5.829 |
| Best Combination | B2M/PPIA | 1.005 | B2M/PPIA | 0.007 | B2M/PPIA | 0.460 |
For GeNorm, lower stability values (M) indicate greater stability. In the case of NormFinder, stability is calculated from inter- and intra-group variation. By grouping the tissues into tumour and normal the best combination of genes was identified. For geNorm stability was based on the estimation of pair-wise variation. QBasePlus through its components, geNorm and qBase, identified coefficient of variation (CV) and stability (M) values and thereby the best combination of genes for normalisation only when more than one gene is used.
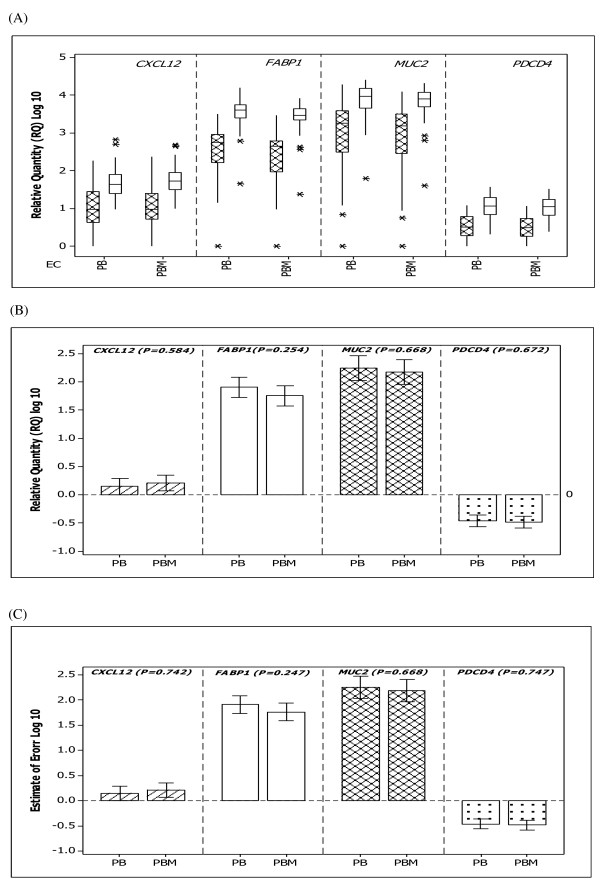
Figure 3.
Relative quantity of CXCL12, FABP1, MUC2 and PDCD4 in colorectal tissue. Error bars indicate 95% confidence intervals. No significant differences in the relative quantities of target genes were found using a combination of PPIA and B2M (PB) genes in comparison to the use of combination of PPIA, B2M and MRPL19 (PBM) EC genes (ANOVA).
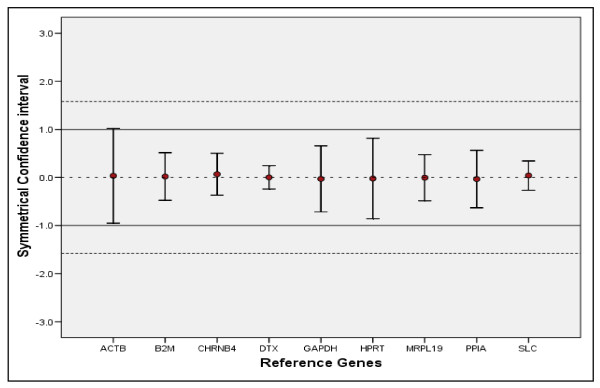
Figure 4.
Equivalence test for candidate control genes in colorectal tissue. Differences in logarithmic expression levels between tumour and normal tissues (●) are indicated. The upper and lower bars of each line indicate the upper and lower limits of the symmetrical confidence intervals, respectively. The deviation area (-1, 1) for a fold change of 2 or less is plotted as a continuous line while the deviation area of (-1.58, 1.58) for a fold change of 3 is plotted as a dotted line.
Association between EC genes and target genes
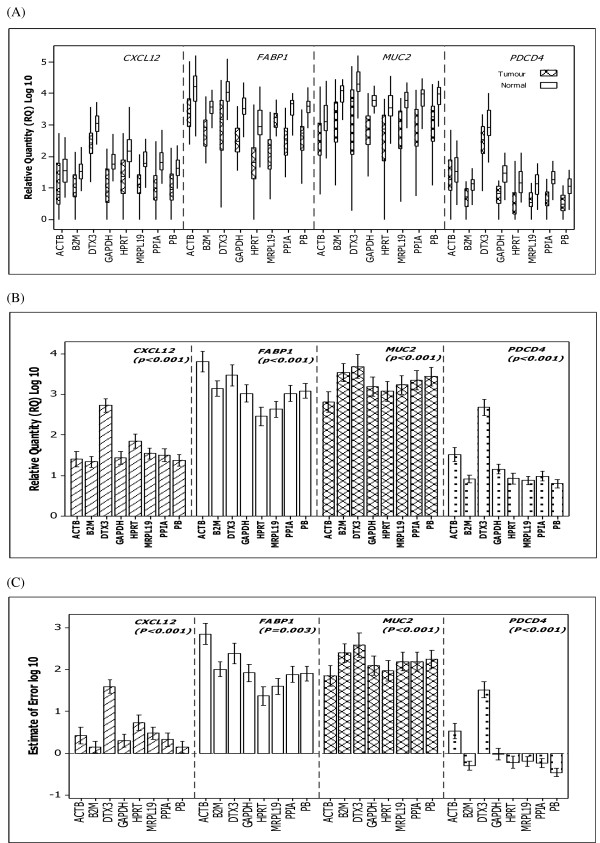
There was a significant effect of the expression of the candidate EC genes on relative expression of CXCL12 (p < 0.001), FABP1 (p < 0.001), MUC2 (p < 0.001) and PDCD4 (p < 0.001) (figure 5A and 5B). Moreover, a significant effect of the choice of EC with regard to the estimation of error (figure 5C) was also detected. These findings were further confirmed for each EC gene compared to each other by ANOVA Tukey post hoc tests (Additional files 1). The combined use of B2M and PPIA significantly reduced the magnitude of error in comparison to the use of either gene individually for both CXCL12 and PDCD4 expression. The addition of a third EC gene (MRPL19) to the B2M/PPIA combination did not result in any further improvement of the estimation of error (figure 3C).
Figure 5.
Relative quantity of target gene expression in colorectal tissues relative to each EC gene and to the geometric mean of the combined use of PPIA and B2M (PB). (A) Target gene expression in tumour versus normal using either individual candidate EC genes or the PB combination. (B) Significant differences in relative gene expression values as determined using ANOVA to compare mean expression levels across all tissues using either individual EC genes or PB in combination. (C) One way ANOVA indicating a reduction in the magnitude of error when the PB combination was used to normalise expression of CXCL12 (p < 0.001) and PDCD4 (p < 0.001) in comparison to the use of individual EC genes. See Table 1 Additional files for Post Hoc tests. Error bars indicate 95% confidence intervals.
Non-normalised expression levels of target genes
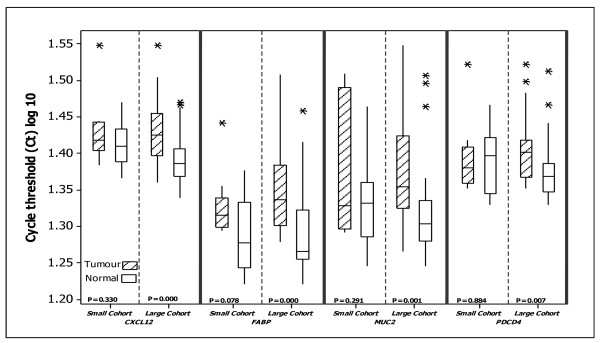
To assess whether normalisation was necessary in a large cohort such as this in which the biological effect of the target genes is already established, we compared the expression of the four target genes in tumour vs. normal tissues using non-normalised cycle threshold (Ct) values in the entire sample set (n = 64) and in a sub-set of randomly selected 10 normal and 10 tumour tissues (n = 20). This analysis showed down-regulated target gene (CXCL12, FABP1, MUC2 and PDCD4) expression in tumour compared to normal tissues (figure 6), in keeping with their documented tumour suppressor functions, when using the larger set of samples. The unchanged target gene expression levels in the large cohort could be explained by the fact that in larger sample sizes the biological milieu may diminish subtle variations in individual samples. In contrast, when the smaller sample size was used, no significant differences in target gene expression were observed. Furthermore the expression levels of PDCD4 appeared slightly higher in tumours than in normal tissues. When the same subset of 20 samples were normalised with PPIA/B2M, significant differences in target gene expression were observed.
Figure 6.
Non-normalised cycle threshold (Ct) of CXCL12, FABP1, MUC2 and PDCD4 in colorectal tissue. Using this approach, the expression of each gene appears to be down-regulated in tumours compared to normal tissues in the large cohort of patients (30 tumour and 34 normal tissue specimens), similar to previous published reports of reduced expression in colorectal tumours. No significant differences were noted in expression levels of target genes when using the small cohort of patients (10 tumour and 10 normal tissue specimens) (2-sample t-test). This confirms the effect of sample size on findings when using non-normalised Ct values and therefore the importance of normalisation especially in such type of studies
Discussion
Since its introduction in 1996 [38] many methods have been developed for the analysis real-time quantitative PCR data. Relative quantification has come to the fore as the method of choice due to its superior flexibility and reduction in inherent variation associated with sample preparation. Prior to the availability of high-throughput realtime PCR instrumentation, a handful of genes were commonly used to normalise real-time data. Major technological advances enabling high throughput analysis of both samples and target genes have enabled investigation and validation of putative EC genes prior to their use to normalise target gene expression. It is now accepted that the use of more than one gene to normalise RQ data improves experimental accuracy compared to the use of a single EC gene [8,35,38]
In their study of EC gene expression in breast and colon cancer tissues Tricarico et al [39] illustrated significant variation in the expression levels of 10 commonly used housekeeping genes including 18S rRNA, both between individuals and between biopsies taken from the same patient. They concluded that normalisation to a single EC gene was inappropriate for human tissue samples. Moreover, Vandesompele et al identified errors of up to 6.5 fold when a single EC gene was used in comparison to the use of multiple genes for data normalisation [8] thereby clearly indicating the potential for superior accuracy when due consideration is paid to the choice of EC genes.
Many analytical programmes for relative quantification have been developed, certain of which enable the identification of EC genes from a study population [37,40,41]. In the present study the stability of expression of candidate EC genes was determined using a pair-wise comparison model: geNorm [8] and an MS Excel ANOVA based model, NormFinder [35]. No effect of disease status EC gene expression was identified in colorectal tissue. Since both geNorm and NormFinder are based on the assumption that candidate genes are not differentially expressed between samples, this was an important first step prior to their continued use [9,10].
In this study GeNorm was used to identify the most stably expressed EC genes from our panel of candidates and also provided a measure of the optimal number of EC genes. B2M and PPIA were identified as the most stable pairing. In order to achieve a pair-wise variation value (V) below the cut-off of 0.15 additional genes should theoretically be used; however this cut-off point is not absolute [14] and may not always be achievable [42]. No significant difference in target gene expression was observed when the top three most stable EC genes identified by geNorm were used confirming that using of a pair of genes may be more practicable given cost, work load and sample availability considerations.
NormFinder was designed to identify EC genes with the lowest stability values; these values are calculated based on intra- and inter-group variation. In this study NormFinder was used to define the best combination of genes using tumour and normal as group identifiers in the calculations. MRPL19 was selected as the most stable gene using these criteria; however B2M and PPIA were highlighted as the best combination of genes with even lower stability value compared to MRPL19 alone. QBasePlus real-time PCR data manager programme was developed based on geNorm and qBase [36] algorithms. QBasePlus was used to confirm our selection of the B2M and PPIA pairing as the best combination of ECs in colorectal tissue.
Equivalence testing was developed in biostatistics to address the situation where the aim is not to show the difference between groups, but rather to establish that two methods are equal to one another. In equivalence testing, the null hypothesis is that the two groups are not equivalent to one another, and hence rejection of the null indicates that the two groups are equivalent. Therefore, as stated by Haller et al, there is a risk of accepting non-differentially expressed genes as suitable controls although they are not equivalently expressed [43]. Equivalence of expression between tumour and normal colorectal tissue was confirmed for all candidate EC genes using the equivalence test and a fold cut-off of 2. DTX3, B2M, MRPL19 and PPIA showed the minimum of variability in the confidence interval hence can be used for normalisation.
In their study to identify EC genes to monitor enterocyte differentiation and to compare normal and adenocarcinoma of the colon from microarray data, Dydensborg et al [5] recommended RPLP0 for normalising gene quantification in human intestinal epithelial cells and B2M for studying gene expression in human colon cancer. In addition, Blanquicett et al [44] analysed the extent of variability in gene expression between tumour and normal colorectal and liver tissues using two-tailed T tests. They showed that 18S, S9 and GUS were the least variable genes in normal and metastatic liver specimens and were also appropriate for normal and tumour colorectal tissues. In the present study, we confirmed that more than one EC gene is required for optimal normalisation in colorectal tissue. We used clinico-pathologically diverse tissues to systematically evaluate normalisation of gene expression data in colorectal tissues. We also conducted equivalence testing to confirm the equality of expression of each EC gene. Thereby, the risks of incorrect rejection (type 1 error) and of false negativity (type 2 error) were minimised.
As stated above significant differences in target gene expression were noticed when using each of the EC genes and the combination of PPIA and B2M. Moreover, significant effect of EC on the magnitude of error associated with estimation of target gene expression was also determined in this study (figure 6). Our results were further confirmed by post hoc testing of individual levels of EC gene expression (Additional files 1). Reduction in the magnitude of error achieved using the combination of PPIA and B2M in comparison to using individual EC genes alone, further indicates that using two EC genes to normalise real-time data achieves greater accuracy in the determination of gene expression levels.
Conclusions
The findings reported in this study confirm that use of two EC genes to normalise RQ-PCR data resulted in superior accuracy in the quantification of gene expression in colorectal tissue. The combined use of B2M and PPIA was validated as the optimal pair of EC genes with which to estimate the expression of all four target genes in colorectal cancer tissue. Although these ECs may not be ideal in other tissue types, the approach described herein could serve as a template to identify valid ECs in other tissue types.
Methods
Tissue Samples
A study group of 64 biopsies of human colon tissue samples was gathered from consenting patients at the time of primary curative surgical resection at Galway University Hospital, Ireland. The cohort comprised of 30 colorectal tumour specimens and 34 and tumour-associated normal (TAN) tissues. Following excision, all samples were subject to histopathological review prior immediate snap-freezing in liquid nitrogen and archival at -80°C until further use. Concomitant clinicopathological data on patients and specimens was obtained from the Department of Surgery Biobank, NUI Galway as detailed in Table 4. Ethical approval for this study was granted by the Clinical Research Ethics Committee, Galway University Hospitals.
Table 4.
Candidate endogenous control (EC) genes and their PCR amplification efficiencies (E)
| EC | Function | Chromosomal Location | Amplicon Size (bp) | Assay Identifier* | E (%) |
|---|---|---|---|---|---|
| B2M | Defence/immunity | 15q21-22.2 | 64 | Hs00187842_m1 | 101.8 |
| GAPDH | Oxidoreductase, dehydrogenase | 12p13 | 122 | Hs99999905_m1 | 99.8 |
| PPIA | Isomerase | 7p13 | 98 | Hs99999904_m1 | 96.6 |
| HPRT | Glycosyl transferase | Xq26.1 | 100 | Hs99999909_m1 | 97.9 |
| MRPL19 | Protein biosynthesis | 2q11.1-11.2 | 72 | Hs00608519_m1 | 102.2 |
| ACTB | Cytoskeletal structure | 7p15-12 | 171 | Hs99999903_m1 | 95.2 |
| DTX3 | Signals transduction | 12q13.3 | 64 | Hs00400987_m1 | 99.1 |
| SLC25A23 | Mitochondrial carrier | 19p13.3 | 86 | Hs00225469_m1 | 97.8 |
| CHRNB4 | Nicotinic receptor | 15q24 | 75 | Hs00609523_m1 | 103.6 |
| RTDR1 | Aminopeptidase transport | 22q11.2 | 112 | Hs00205353_m1 | UD |
| HCRT | Homeostatic regulator | 12q21 | 101 | Hs00533664_m1 | UD |
| APOC4 | Apo-lipoprotein | 19q.2 | 144 | Hs00155791_m1 | UD |
| KRTAP12-3 | Acetylgalactoa-minyltransferase | 3q25 | 83 | Hs01651247_s1 | UD |
* Applied Biosystems TaqMan® gene expression assay ID
UD: undetermined
RNA Extraction and Analysis
Tissue samples (50-100 mg) were homogenised using a hand-held homogenizer (Polytron® PT1600E, Kinematica AG, Littau-Luzem, Switzerland) in 1-2 ml of QIAzol reagent (Qiagen, Crawley, UK). To minimise variation in sample processing, tumour and TAN samples were homogenised separately, but on the same day. RNA was extracted as previously described (Davoren et al) using the RNeasy® Plus Mini Kit and RNeasy MinElute® cleanup kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. Briefly, large (> 200 nt) and small RNA (< 200 nt) fractions were isolated separately. For this study, only large RNA was utilised for further analysis. RNA was eluted in 60 μl volumes and stored at -80°C.
RNA concentration and purity was assessed in duplicate samples using a using a NanoDrop™ ND-1000 Spectrophotometer (Thermo Fisher Scientific, USA). RNA integrity was evaluated using the RNA 6000 Nano Chip Kit (Series II) and the Agilent 2100 Bioanalyzer System (Agilent technologies, Palo Alto, CA, USA). An RNA integrity number (RIN) was generated for each sample using the Agilent 2100 Expert Software (Version B.02.03) based on the ratio of ribosomal bands and also the presence or absence of degradation products on the electrophoretic and gel-like images. A threshold value of RIN ≥ 7 was applied and RNA purity was verified by an average A260/A280 ratio of 1.98 (range 1.97-2.01) and A260/A230 ration of 1.7 (range 1.5-1.83).
Candidate Endogenous Control Genes
Based on literature search six commonly used candidate endogenous control genes were selected for analysis: ACTB, GAPDH, HPRT, B2M, PPIA and MRPL19. An additional panel of seven genes: HCRT, SLC25A23, DTX3, APOC4, RTDR1, KRTAP12-3 and CHRNB4, was also selected for analysis (Table 2). To our knowledge all genes have independent cellular functions and were assumed not to be co-regulated.
cDNA Synthesis and RQ-PCR
First strand cDNA was synthesised using Superscript™ III reverse transcriptase (Invitrogen Life technologies, Paisley, UK) and random primers (N9; 1 μg, MWG Biotech, AG, Ebersberg, Germany). Negative control samples were included in each set of reactions. Reactions were incubated at 25°C for 5 minutes followed by 50°C for 1 hour and final denaturation at 72°C for 15 minutes. Samples were subsequently diluted to 50 μL in nuclease-free water and stored at -20°C. The expression of each EC gene was analysed by RQ-PCR using TaqMan® gene expression assays using a 7900HT instrument (Applied Biosystems, Foster city, USA). All reactions were performed in 20 μL reactions, in triplicate within the same PCR run. Negative controls were included for each gene target under assay. On each plate, an interassay control was included to account for any variations between runs. For each well 2 μl of cDNA from each sample was added to 18 μl of PCR reaction mix which consisted of 10× TaqMan® universal master mix, No AmpErase UNG, 7× nuclease free water and 1× gene expression assay primer-probe mix (Applied Biosystems, Foster city, USA). The PCR reactions were initiated with a 10 minute incubation at 95°C followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds, in accordance with the manufacturer's recommendations.
PCR Amplification Efficiency
Amplification efficiencies for each EC gene assay were calculated applying the formula E = (10-1/slope - 1) × 100, using the slope of the plot of Ct versus log input of cDNA (10-fold dilution series). A threshold of 10% above and below 100% efficiency was applied. PCR amplification efficiency for each candidate EC gene is shown in table 2.
Data Analysis
Cycle threshold (Ct) is defined as the PCR cycle number at which the fluorescence generated from amplification of the target gene within a sample increases to a threshold value of 10 times the standard deviation of the base line emission and is inversely proportionate to the starting amount of the target cDNA. QBasePlus was used for calculation of PDCD4 expression relative to each of the EC genes. It applies ΔΔCt method was used where ΔΔCt = (Ct target gene, test sample - Ct endogenous control, test sample) - (Ct target gene, calibrator sample - Ct endogenous control, calibrator sample). Relative quantities were corrected for efficiency of amplification and fold change in gene expression between groups was calculated as E-ΔΔCt ± s.e.m. Where more than one endogenous control are used, fold change estimates were calculated using the geometric mean of EC quantities relative to the calibrator sample which could be the minimum, maximum or a named sample or an average.
Stability of the EC genes expression was evaluated with two freely available statistical models, geNorm and NormFinder. It is further validated with qBasePlus. Statistical analysis was carried out with Minitab® 15 (Minitab Ltd, Coventry, UK). Anderson-Darling normality test was applied and parametric tests were used where appropriate. The equivalence test was used to assess the equivalently of expression of the candidate genes between tumour and normal tissues. One-way ANOVA, two-sample t-test, Levene's test and Spearman and Pearson correlations were used to determine association and comparisons between groups. P values < 0.05 were considered statistically significant.
Authors' contributions
EAHK performed the experiments, was responsible for data analyses and drafted the manuscript. KHC contributed to sample preparation from clinical samples and collation of clinicopathological data. NM conceived, designed and supervised experimental work and manuscript editing. JN contributed to statistical analysis of clinical data and drafting of the manuscript. MJK contributed throughout the experiment, critically reviewed the manuscript and participated clinically. All authors read and approved the final manuscript.
Supplementary Material
Table 1 Supplementary data. Post hoc testing of individual levels of EC gene expression.
Contributor Information
Elrasheid AH Kheirelseid, Email: rashmed1111@gmail.com.
Kah Hoong Chang, Email: kahhoong_chang@yahoo.co.uk.
John Newell, Email: john.newell@nuigalway.ie.
Michael J Kerin, Email: michael.kerin@nuigalway.ie.
Nicola Miller, Email: nicola.miller@nuigalway.ie.
Acknowledgements
The authors would like to acknowledge the National Breast Cancer Research Institute (NBCRI) for their continued financial support. We gratefully acknowledge Ms. Emer Hennessy and Ms. Catherine Curran for continued technical assistance.
References
- O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96(19):1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85(1):154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes. 2005;19(2):101–109. doi: 10.1016/j.mcp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Dydensborg AB, Herring E, Auclair J, Tremblay E, Beaulieu JF. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am J Physiol Gastrointest Liver Physiol. 2006;290(5):1067–1074. doi: 10.1152/ajpgi.00234.2005. [DOI] [PubMed] [Google Scholar]
- Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. BioTechniques. 2005;5(2):209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill RE, Miller N, Kerin MJ. Evaluation and validation of candidate endogenous control genes for real-time quantitative PCR studies of breast cancer. BMC Mol Biol. 2007;8:107. doi: 10.1186/1471-2199-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Molecular Biology. 2008;9:76. doi: 10.1186/1471-2199-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer. 2008;8:20. doi: 10.1186/1471-2407-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviozzi S, Cordero F, Lo Iacono M, Novello S, Scagliotti GV, Calogero RA. Selection of suitable reference genes for accurate normalization of gene expression profile studies in non-small cell lung cancer. BMC Cancer. 2006;6:200. doi: 10.1186/1471-2407-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Ramankulov A, Roigas J, Johannsen M, Ringsdorf M, Kristiansen G, Jung K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol Biol. 2007;8:47. doi: 10.1186/1471-2199-8-47. 2007, 8: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson DT, Brockbank S, Quinn JG, Murphy S, Ravid R, Irvine GB, Johnston JA. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol Biol. 2008;9:46. doi: 10.1186/1471-2199-9-46. 2007, 8: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicinnati VR, Shen Q, Sotiropoulos GC, Radtke A, Gerken G, Beckebaum S. Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer. 2008;8:350. doi: 10.1186/1471-2407-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Simons J. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun. 1999;259(3):523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]
- Valenti MT, Bertoldo F, Dalle Carbonare L, Azzarello G, Zenari S, Zanatta M, Balducci E, Vinante O, Lo Cascio V. The effect of bisphosphonates on gene expression: GAPDH as a housekeeping or a new target gene? BMC Cancer. 2006;6:49. doi: 10.1186/1471-2407-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunth K, Höfler H, Atkinson MJ. Quantification of messenger RNA expression in tumors: which standard should be used for best RNA normalization? Verh Dtsch Ges Pathol. 1994;78:226–230. [PubMed] [Google Scholar]
- Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295(1):17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- Oikarinen A, Mäkelä J, Vuorio T, Vuorio E. Comparison on collagen gene expression in the developing chick emberyotendon and heart. Tissue and development tissue-dependent action of dexamethasone. Biochim Biophys Acta. 1991;1089(1):40–46. doi: 10.1016/0167-4781(91)90082-w. [DOI] [PubMed] [Google Scholar]
- Fedchenko V, Globa A, Kaloshin A, Kapitsa I, Nerobkova L, Val'dman E, Buneeva O, Glover V, Medvedev A. The effect of short-term administration of (-)-deprenyl and isatin on the expressions of some genes in the mouse brain cortex. Med Sci Monit. 2008;14(12):BR269–273. [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46(1-2):69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Wu YY, Rees JL. Variation in epidermal housekeeping gene expression in different pathological states. Acta Derm Venereol. 2000;80(1):2–3. doi: 10.1080/000155500750012397. [DOI] [PubMed] [Google Scholar]
- Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM. Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. Journal of Neurochemistry. 1996;66(3):928–935. doi: 10.1046/j.1471-4159.1996.66030928.x. [DOI] [PubMed] [Google Scholar]
- Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259(5093):365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29(1):23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2-3):95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Foss DL, Baarsch MJ, Murtaugh MP. Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and beta-actin mRNA expression in porcine immune cells and tissues. Animal Biotechnology. 1998;9(1):67–78. doi: 10.1080/10495399809525893. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29(2):332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Wei Q, Guan Y, Cheng L, Radinsky R, Bar-Eli M, Tsan R, Li L, Legerski RJ. Expression of five selected human mismatch repair genes simultaneously detected in normal and cancer cell lines by a nonradioactive multiplex reverse transcription-polymerase chain reaction. Pathobiology. 1997;65(6):293–300. doi: 10.1159/000164141. [DOI] [PubMed] [Google Scholar]
- Akishima-Fukasawa Y, Nakanishi Y, Ino Y, Moriya Y, Kanai Y, S H. Prognostic significance of CXCL12 expression in patients with colorectal carcinoma. Am J Clin Pathol. 2009;132(2):202–210. doi: 10.1309/AJCPK35VZJEWCUTL. [DOI] [PubMed] [Google Scholar]
- Lawrie LC, Dundas SR, Curran S, GI M. Liver fatty acid binding protein expression in colorectal neoplasia. Br J Cancer. 2004;90(10):1955–1960. doi: 10.1038/sj.bjc.6601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Popova NV, Yang WC, Lozonschi I, Tadesse S, Kent S, Bancroft L, Matise I, Cormier RT, Scherer SJ. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res. 2008;68(18):7313–7322. doi: 10.1158/0008-5472.CAN-08-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun Z, Yang HS. Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene. 2008;27(11):1527–1535. doi: 10.1038/sj.onc.1210793. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller F, Kulle B, Schwager S, Gunawan B, von Heydebreck A, Sultmann H, Fuzesi L. Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalization. Anal Biochem. 2004;335(1):1–9. doi: 10.1016/j.ab.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes and immunity. 2005;6(4):279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309(2):293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Szabo A, Perou CM, Karaca M, Perreard L, Quackenbush JF, Bernard PS. Statistical modeling for selecting housekeeper genes. Genome Biol. 2004;5(8):R59. doi: 10.1186/gb-2004-5-8-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellek S. Testing statistical hypothesis of equivalence. London: Chapman & Hall; 2003. [Google Scholar]
- Blanquicett C, Johnson MR, Heslin M, Diasio RB. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. Anal Biochem. 2002;303(2):209–214. doi: 10.1006/abio.2001.5570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 Supplementary data. Post hoc testing of individual levels of EC gene expression.