Abstract
Proteolyzed peptides provide the basis for mass-analyzed hydrogen/deuterium exchange (HDX) for mapping solvent access to various segments of solution-phase proteins. Aspergillus saitoi protease type XIII and porcine pepsin can generate peptides of overlapping sequences and high sequence coverage. However, if disulfide bonds are present, proteolysis can be severely limited, particularly in the vicinity of the disulfide linkage(s). Disulfide bonds cannot be reduced before or during the H/D exchange reaction without affecting the protein higher-order structure. Here, we demonstrate simultaneous quench/digestion/reduction following H/D exchange, for subsequent mass analysis. Proteolysis is conducted in the presence of TCEP·HCl and urea, and all other steps of the H/D exchange and analysis are maintained. This method yields dramatically increased sequence coverage and localization of solvent exposed segments for mass-analyzed solution-phase H/D exchange of proteins containing disulfide bonds.
Keywords: Fourier transform, ion cyclotron resonance, FTICR, FTMS, H/D exchange
INTRODUCTION
Hydrogen/deuterium exchange (HDX) coupled with mass spectrometry is widely used to map solvent exposure for various segments of a protein.1–5 Localization of the site(s) of H/D exchange is achieved by digesting the protein or protein complex by an acid-active protease, followed by rapid (to minimize back-exchange of D to H) liquid chromatography desalting/separation and determination of the deuterium uptake for each peptide by (typically) electrospray ionization (ESI) mass spectrometry (MALDI MS may also be used as the detection method6). The protein-ligand binding surface and/or conformational changes induced on binding may be characterized by comparing free protein and protein-ligand complex. Recent applications of HDX MS to study protein conformational changes include drug resistance mechanism,7 allosteric regulation for signal transduction pathways,8 Abl kinase conformational change upon mutation and degradation,9 and virus scaffolding protein assembly.10, 11
High sequence coverage for the proteolyzed peptides is clearly essential. Pepsin is commonly used to digest a protein for HDX MS, due to its activity under acidic conditions and its broad specificity to generate peptides of reasonably small length (typically 10–20 amino acids). Recently, Cravello et al. introduced acid-active protease type XIII from Aspergillus saitoi and protease type XVIII from Rhizhopus to increase sequence coverage.12 In our experience, protease type XIII yields higher sequence coverage than pepsin.13 Moreover, like trypsin, protease type XIII prefers to cleave at the C-terminus of basic amino acids (Arg, Lys and His), for improved electrospray ionization efficiency (because basic amino acids provide a positive charge), and thus higher mass spectral signal/noise ratio. In addition, protease type XIII also exhibits much less self-digestion than pepsin, and thus fewer interfering peptides in the eventual mass spectrum.
Many proteins (especially larger ones) have disulfide bonds that are important for folding and stability,14 e.g., secretary proteins, lysosomal proteins, and extracellular portion(s) of membrane proteins. However, disulfide linkage(s) can render extensive segments of the protein inaccessible to proteolysis, and the available proteolytic segments are too long to allow localization of the site(s) of H/D exchange. Therefore, the disulfide bonds need to be reduced for effective HDX MS analysis.15–18 For example, Kattat et al. reduced intact chicken egg lysozyme by adding dithiothreitol (DTT) to the protein in H2O,15 followed by H/D exchange for both the reduced and non-reduced proteins.
However, if HDX is to probe the native conformation of a protein or complex, reduction cannot be performed before or during the H/D exchange reaction, because the disulfide bond reduction can change the protein conformation. Therefore, Yan et al.16 reduced the disulfide bonds after H/D exchange under quench conditions (0 °C, pH 2.5) by tris-(2-carboxyethyl) phosphine hydrochloride (TCEP·HCl) (producing an acidic condition at pH ~2.5) and followed with immobilized pepsin column digestion. Li et al.18 also used TCEP·HCl to quench the H/D exchange reaction and reduce the peptide disulfide bonds. Because this experiment was for peptides rather than proteins, no proteolytic digestion was used.
To the best of our knowledge, no one has reported simultaneous protein reduction and proteolysis for solution-phase HDX experiments. In this short paper, we introduce a novel simultaneous quench, reduction, and digestion method to simultanously stop the H/D exchange reaction, and unfold, reduce, and digest a protein containing disulfide bonds simultaneously after the HDX reaction is complete. This protocol provides high sequence coverage spanning cysteine-containing segments, without affecting the rest of the automated experimental procedure for solution-phase HDX MS. This method has potential to greatly expand the application of HDX MS to characterize conformation and ligand-binding effects in large, disulfide-containing proteins.
EXPERIMENTAL PROCEDURES
Materials
Protease type XIII from Aspergillus saitoi, pepsin from porcine stomach, HEPES, sodium chloride, deuterium oxide (min. 99.96% atom% D), Tris (2-carboxyethyl)phosphine hydrochloride (TCEP·HCl), urea, and formic acid were purchased from Sigma Aldrich (St. Louis, MO). HPLC grade H2O and acetonitrile were purchased from VWR International (Suwanee, GA). Urea was purchased from Fisher Scientific (Suwanee, GA).
Digestion
Various digestion conditions (with or without reducing and denaturing agents) were tested for myoglobin (Sigma Aldrich (St. Louis, MO)), hepatitis-C virus (HCV) proteins (wild-type and mutant) from Hengli Tang’s lab at Florida State University, and sRAGE proteins (expressed from BacMam and E. coli from Abbott (see Results and Discussion).
Hydrogen/Deuterium Exchange
HDX experiments for sRAGE proteins were performed as described previously.19 Our entire HDX experiment is automated by a Leap robot (HTS PAL; Leap Technologies, Carrboro, NC), to perform sample dilution (initiation of the exchange reaction), reaction time control, quench, digestion time control, and injection onto the LC column. All of the experiments were performed in triplicate to increase reproducibility.
Briefly, H/D exchange was initiated by diluting 5 μL of ~25 μM sRAGE stock protein into 45 μL of 25 mM HEPES and 100 mM NaCl buffer in D2O (pH meter reading = 7.0). For a blank control, the dilution was made in H2O-based buffer with the same concentrations of HEPES and NaCl. H/D exchange was allowed to proceed for 0.5, 1, 2, 4, 8, 15, 30, 60, 120, and 240 min prior to simultaneous quench, proteolysis and reduction. The controls and the timed reactions were performed in triplicate. Each HDX reaction was quenched by addition of protease solution at 1:1 (v/v) to decrease the final pH to ~2.3 to ~2.5, followed by 2 min protease digestion and injection of the digested sample (through a 30 μL sample loop) to a liquid chromatograph. The entire HDX was performed at 0.4 °C controlled by a Huber recirculating H2O bath (Peter Huber, Germany).
Simultaneous Digestion/Reduction
The sRAGE protein was digested with either pepsin (~1.4 mg/mL) or protease type XIII (~1.2 mg/mL) as described previously.19 To reduce the disulfide bonds in the proteins and provide higher sequence coverage, concentrations were optimized in the final solution as follows: 3 fold diluted protease type XIII (~4 mg/mL) or pepsin (0.5 mg/mL) mixed with 100 mM TCEP·HCl and 4 M urea. In the final quench/digestion/reduction solution, the pH meter reading was 2.52, temperature = 0.4 °C. Stability of the protease activity was also tested for that mixture.
LC Separation, Mass Spectrometry and Data Analysis
A Jasco HPLC/SFC instrument (Jasco, Easton, MD) was interfaced to the LEAP robot to desalt and separate the peptides after proteolysis. We used a fast LC separation to minimize back exchange 19. The digested peptides were desalted/separated by a Pro-Zap C18 column (Grace Davidson, Deerfield, IL) with a 1.5 min fast gradient (2% B to 95% B in 1.5 min at 300 μL/min, A: acetonitrile/H2O/formic acid, 5/94.5/0.5; B: acetonitrile/H2O/formic acid, 95/4.5/0.5). The peptide-containing eluent was introduced to a custom-built 14.5 T FT-ICR mass spectrometer20 with a Thermo Fisher Scientific linear quadrupole ion trap (LTQ) front end at ~400–500 nL/min for efficient microelectrospray ionization21 and accurate mass measurement. Mass spectra were recorded from m/z 400–2000 at high mass resolving power (m/Δm50%=200,000 at m/z 400, in which Δm50% is magnitude-mode mass spectral peak full width at half-maximum height). The total data acquisition period for each sample was 6 min. External ion accumulation22 was performed in the linear ion trap with a target ion population of 1 million charges collected for each FT-ICR measurement. LTQ-accumulated ions were transferred (~1 ms transfer period) through three octopole ion guides (2.2 MHz, 250 Vp-p) to a capacitively coupled23 closed cylindrical ICR cell (55 mm i.d.) for analysis. Automatic gain control24 and high magnetic field25 provided excellent external calibration mass accuracy (typically better than 500 ppb rms). Data were collected with Xcalibur software (ThermoFisher) and analyzed by a custom analysis package,26 permitting reliable identification of the peptides and accurate assignment of the extent of deuterium incorporation.
RESULTS AND DISCUSSION
Protease Activity in the Presence of Denaturing and Reducing Agents
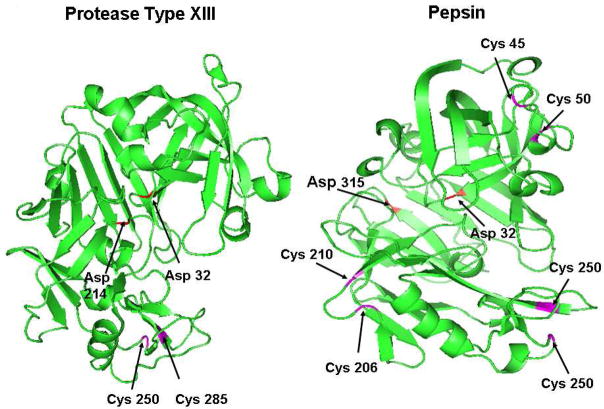
For this approach to succeed, the protease must maintain its activity in the presence of the reducing agent. It is therefore useful to note the number and sequence location(s) of disulfide bonds in protease type XIII and pepsin. Figure 1 shows the X-ray crystal structures for protease type XIII and pepsin, with the disulfide bonds highlighted in red. Protease type XIII contains only one disulfide bond, and it is distant from its active site in the catalytic domain. Thus, reduction of this disulfide bond should not significantly affect its proteolytic acitivity. In contrast, mature pepsin contains 6 cysteines linked by three disulfide bonds;27 thus, disulfide reduction would be expected to reduce its catalytic efficiency. We therefore tested the protease type XIII and pepsin activity against a standard protein: horse heart myoglobin.
Figure 1.
Three dimensional x-ray crystal structures of protease type XIII (PDB ID: 1IBQ) and pepsin (PDB ID: 5PEP). Cysteine residues forming one disulfide bond for protease type XIII and three disulfide bonds for pepsin are highlighted in magenta; Asp 32 and Asp 214 for protease type XIII; and Asp 32 and Asp 315 for pepsin in the active sites of the proteases are highlighted in red.
As previously noted,13 protease type XIII generates higher sequence coverage, with more fragments and shorter average peptide length than pepsin (see Table 1). Here we also list the digestion results of myoglobin by protease type XIII and pepsin in the presence of a denaturant (urea) and the reducing agent (TCEP·HCl). The protease solution was prepared just before digestion by mixing protease type XIII with TCEP·HCl and urea. As seen in Table 1, protease type XIII maintained 100% sequence coverage, and generated 3 fewer fragments (65 vs. 68). Proteolyzed peptide average length was a little longer (12 vs. 11). For pepsin, the sequence coverage was also maintained at 98%, but the fragment number was 22 fewer (33 vs. 54) and peptide average length increased from 15 to 17. Thus, the presence of denaturing and reducing agents had more effect on pepsin than on protease type XIII.
Table 1.
Proteolysis results for myoglobin digested by Aspergillus saitoi protease type XIII or porcine pepsin without or with TCEP/urea added.
| Sequence Coverage | Number of Fragments | Average Length | |
|---|---|---|---|
| Myoglobin + Protease XIII | 100% | 68 | 11 |
| Myoglobin + Protease XIII/TCEP/urea | 100% | 65 | 12 |
| Myoglobin + Pepsin | 98% | 54 | 15 |
| Myoglobin + Pepsin/TCEP/urea | 98% | 32 | 17 |
Temporal Stability of the Proteases in the Presence of Denaturing and Reducing Agents
To maintain our present automation settings for the experiment, it is preferable to prepare the protease stock solution (including denaturing and reducing agents) without changing the remaining procedures, provided that the proteases maintain activity in the presence of denaturing and reducing agents. We therefore tested protease activity 1, 2 and 4 hours after preparation of the protease solution (see Table 2). For both proteases, digestion activity was not significantly reduced by incubation in TCEP·HCl and urea. Therefore, no extra steps are needed to automate the experiment.
Table 2.
Proteolysis results for myoglobin digested by proteases in the presence of TCEP/urea, after 1, 2, and 4 hours of preparation.
| Time from Preparation (h) | Sequence Coverage | Number of Fragments | Average Length | |
|---|---|---|---|---|
| Protease XIII/TCEP/urea | 1 | 100% | 56 | 13 |
| Protease XIII/TCEP/urea | 2 | 100% | 57 | 13 |
| Protease XIII/TCEP/urea | 4 | 100% | 54 | 13 |
| Pepsin/TCEP/urea | 1 | 98% | 31 | 16 |
| Pepsin/TCEP/urea | 2 | 98% | 30 | 16 |
| Pepsin/TCEP/urea | 4 | 98% | 33 | 17 |
Increased Sequence Coverage for Hepatitis C Virus (HCV) NS5A Domain II
HCV NS5A is a multi-domain protein that is critical for the life cycle of this human virus. Domain II of NS5A is natively unfolded28 and mediates protein-protein interactions that are important for both viral replication and pathogenesis.29 This domain has 102 amino acids and one disulfide bond for both the wild-type and the DEYN (Asp71 to Glu71 and Tyr72 to Asn 72 mutations) mutant proteins. The DEYN mutations alter the conformation of the domain and confer to HCV the ability to replicate independent of cyclophilin A,(Yang, Grise and Tang, unpublished data) a peptidylprolyl isomerase essential for the virus to replicate.30 Table 3 shows the sequence coverage for the WT and mutant HCV proteins by protease type XIII and pepsin, without and with TCEP/urea. With TCEP/urea added, sequence coverage by protease type XIII reached 100% for both the wild-type and mutant proteins. Note that the average peptide is a little longer with TCEP/urea added than for the protease alone, with the caveat that the protease alone did not even cover the regions near Cys (sequence coverage is only 64% and 67% for the wild-type and the mutant proteins). For pepsin, sequence coverage also improved (to more than 90%) with TCEP/urea added, but not as good as for protease type XIII, and the average proteolytic peptides were longer.
Table 3.
Proteolysis results for hepatitis-C virus wild-type and mutant proteins digested by protease type XIII or pepsin, without or with TCEP/urea added.
| Sequence Coverage | Number of Fragments | Average Length | |
|---|---|---|---|
| HCV Wild-Type + Protease XIII | 64% | 45 | 11 |
| HCV Wild-Type + Protease XIII/TCEP/urea | 100% | 41 | 12 |
| HCV Wild-Type + Pepsin | 64% | 19 | 11 |
| HCV Wild-Type + Pepsin/TCEP/urea | 96% | 28 | 15 |
| HCV Mutant + Protease XIII | 67% | 40 | 12 |
| HCV Mutant + Protease XIII/TCEP/urea | 100% | 36 | 13 |
| HCV Mutant + Pepsin | 64% | 18 | 10 |
| HCV Mutant + Pepsin/TCEP/urea | 90% | 25 | 14 |
Figure 2 illustrates the proteolyzed peptides mapped onto the primary sequence of the wild-type HCV protein. Without TCEP and urea, the segments containing the two Cys forming one disulfide bond are not accessed by either protease type XIII or pepsin. For a mixture of protease type XIII with only TCEP (i.e., no urea) (data not shown), the disulfide bond was not effectively reduced, resulting in low sequence coverage. With incubation in TCEP·HCl and urea, protease type XIII yielded 100% sequence coverage and pepsin yielded 96%, both with the regions adjacent to the two Cys covered. Urea serves to unfold the protein and makes it more accessible to the reducing agent. The peptides generated by pepsin have longer average length, and thus lower sequence resolution. (Urea concentration was limited to 4 M, so as not to significantly denature the proteases themselves.) In conclusion, protease type XIII with TCEP·HCl and urea the best choice for proteolysis, if the sample amount is limited; if enough sample is available, additional digestion with pepsin, TCEP·HCl and urea can further increase sequence resolution.
Figure 2.
Proteolyzed peptides mapped onto the primary sequence of HCV wild-type NS5A domain II by protease type XIII and pepsin, without or with added TCEP/urea.
Increased Sequence Coverage of sRAGE Proteins for HDX MS and Unaffected Back-Exchange Level
We also tested the reduction/digestion method to increase sequence coverage for sRAGE (soluble portion of the receptor of advanced glycation endproducts) proteins from Abbott. sRAGE proteins expressed from either BacMam or E. coli each contain 6 Cys. The proteins were initially digested by protease type XIII and pepsin without any reducing and denaturing agents. However, sequence coverage for the common fragments that provide a basis for deuterium incorporation comparison was not high (33% for pepsin and 48% for protease type XIII). With the addition of reducing and denaturing agents, the common fragments from the two constructs reached 89%, and thus a good basis for comparing their conformations. The digestion patterns for protease type XIII alone and with TCEP/urea are not identical. Combination of the results from different digestion conditions yields higher sequence coverage and better sequence resolution, provided that enough protein is available. Results of digestion by protease type XIII and pepsin for the two sRAGE constructs are listed in Table 4.
Table 4.
Proteolysis results for protease type XIII or pepsin digestion, without or with TCEP/urea, of sRAGE protein expressed from BacMam or E. coli.
| Sequence Coverage | Number of Fragments | Average Length | |
|---|---|---|---|
| BacMam sRAGE + Protease XIII | 83% | 57 | 13 |
| BacMam sRAGE + Protease XIII/TCEP/urea | 94% | 59 | 15 |
| BacMam sRAGE + Pepsin | 71% | 21 | 19 |
| BacMam sRAGE + Pepsin/TCEP/urea | 93% | 42 | 21 |
| E. coli sRAGE + Protease XIII | 70% | 40 | 14 |
| E. coli sRAGE + Protease XIII/TCEP/urea | 92% | 52 | 16 |
| E. coli sRAGE + Pepsin | 68% | 26 | 16 |
| E. coli sRAGE + Pepsin/TCEP/urea | 82% | 40 | 17 |
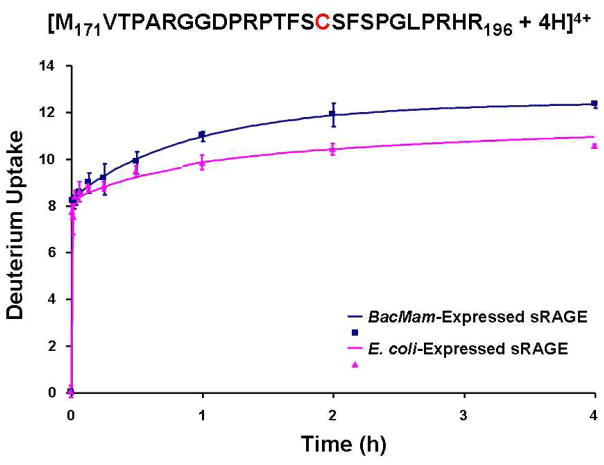
In addition to the digestion test, we also performed a complete H/D exchange MS experiment to compare the deuterium incorporation for the peptides from the two constructs. Figure 3 shows an example of the deuterium uptake curves for a peptide (171–196) containing one Cys residue. The same segment from BacMam-expressed sRAGE has somewhat higher deuterium incorporation than from E. coli-expressed sRAGE. Such differences help to elucidate the effect of glycosylation on sRAGE, a topic for future investigation.
Figure 3.
Deuterium incorporation uptake vs. H/D exchange period for the same proteolytic peptide from sRAGE proteins, with one Cys, expressed from BacMam or E. coli. After H/D exchange, each sRAGE protein was digested by protease type XIII with TCEP/urea added.
Comparison of deuterium incorporation for sRAGE fragments showed no significant difference in the extent of back exchange for the previous digestion method (protease type XIII in 1% FA) and the simultaneous quench/digestion/reduction method, because the pH and temperature of the final solution is similar for both methods (pH 2.52 for the current method vs. pH 2.54 for the prior method). Urea’s tendency to increase pH is counteracted by TCEP·HCl’s decrease in pH.
CONCLUSIONS
To increase proteolytic sequence coverage for proteins with disulfide bonds, we have developed a new protocol to simultaneously quench, reduce, and digest the protein. Proteolytic activity of protease type XIII is not significantly affected in the presence of denaturing and reducing agents, but pepsin suffers somewhat, presumably because protease type XIII has only one disulfide bond (and it is remote from the active site), whereas pepsin has three. Dramatically improved sequence coverage was achieved for the proteins containing disulfide bonds by the proteases in the presence of denaturing and reducing agents, especially for protease type XIII. Both protease type XIII and pepsin are stable in the presence of the reducing and denaturing agents up to more than four hours, and may thus be prepared and used as for the protease alone. No extra experimental steps are needed, and automation is unaffected. Back exchange level is not affected due to unchanged pH and temperature. The present approach promises to extend HDX monitored by mass spectrometry to proteins with disulfide bonds, e.g., extracellular portion of membrane proteins, cell surface receptors, and antibodies.
Acknowledgments
This work was supported by NIH (1R01 GM78359), NSF Division of Materials Research (DMR 0654118), and the State of Florida.
References
- 1.Zhang Z, Smith DL. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engen JR, Smith DL. Anal Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 3.Busenlehner LS, Armstrong RN. Arch Biochem Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Wales TE, Engen JR. Mass Spectrometry Reviews. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 5.Tsutsui Y, Wintrode PL. Curr Med Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 6.Mandell JG, Falick AM, Komives EA. Anal Chem. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 7.Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, English JM, Greig MJ, He YA, Jacques SL, Lunney EA, McTigue M, Molina D, Quenzer T, Wells PA, Yu X, Zhang Y, Zou A, Emmett MR, Marshall AG, Zhang HM, Demetri GD. Proc Nat Acad Sci. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frantom PA, Zhang HM, Emmett MR, Marshall AG, Blanchard JS. Biochemistry. 2009;48:7457–7464. doi: 10.1021/bi900851q. [DOI] [PubMed] [Google Scholar]
- 9.Iacob RE, Dumitrescu TP, Zhang J, Gray NS, Smithgall TE, Engen JR. Proc Natl Acad Sci USA. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE. Nat Struc Mol Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 11.Tuma R, Tsuruta H, French KH, Prevelige PE. J Mol Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cravello L, Lascoux D, Forest E. Rapid Commun Mass Spectrom. 2003;17:2387–2393. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HM, Kazazic S, Schaub TM, Tipton JD, Emmett MR, Marshall AG. Anal Chem. 2008;80:9034–9041. doi: 10.1021/ac801417d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sevier CS, Kaiser CA. Nature Reviews Molecular and Cellular Biology. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 15.Kattat V, Chait BT. J Am Chem Soc. 1993;115:6317–6321. [Google Scholar]
- 16.Yan X, Zhang H, Watson J, Schimerlik MI, Deinzer ML. Protein Sci. 2002;11:2113–2124. doi: 10.1110/ps.0204402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Chou YT, Husain R, Watson JT. Anal Biochem. 2004;331:130–137. doi: 10.1016/j.ab.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Hood RJ, Wedemeyer WJ, Watson JT. Protein Sci. 2005;14:1922–1928. doi: 10.1110/ps.051458905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang HM, Bou-Assaf GM, Emmett MR, Marshall AG. J Am Soc Mass Spectrom. 2009;20:520–524. doi: 10.1016/j.jasms.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG. Anal Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 21.Emmett MRaRMC. J Am Soc Mass Spectrom. 1994;5:605–613. doi: 10.1016/1044-0305(94)85001-1. [DOI] [PubMed] [Google Scholar]
- 22.Senko MW, Hendrickson CL, Emmett MR, Shi SDH, Marshall AG. J Am Soc Mass Spectrom. 1997;8:970–976. [Google Scholar]
- 23.Beu SC, Laude DA. Anal Chem. 1992;64:177–180. [Google Scholar]
- 24.Schwartz JC, Senko MW, Syka JEP. J Am Soc Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 25.Marshall AG, Guan S. Rapid Commun Mass Spectrom. 1996;10:1819–1823. doi: 10.1002/(SICI)1097-0231(199611)10:14<1855::AID-RCM764>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Kazazic S, Zhang H, Schaub TM, Emmett MR, Christopher HL, Blakney GT, Marshall AG. 2009 In submitted. [Google Scholar]
- 27.Cooper JB, Khan G, Taylor G, Tickle IJ, Blundell TL. J Mol Bio. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- 28.Liang Y, Ye H, Kang CB, Yoon HS. Biochemistry. 2007;46:11550. doi: 10.1021/bi700776e. [DOI] [PubMed] [Google Scholar]
- 29.Tang H, Grise H. Clin Sci. 2009;117:49–65. doi: 10.1042/CS20080631. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. J Virology. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]