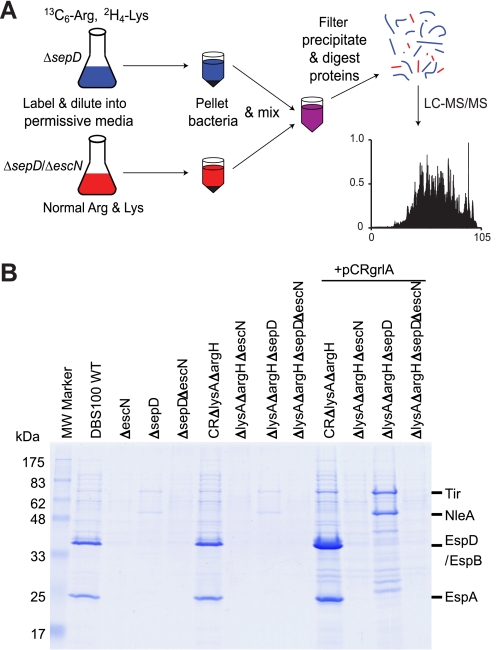
FIGURE 1.
SILAC analysis of the type III secretome by C. rodentium. A, overview of the SILAC protocol using C. rodentium ΔsepD and ΔsepDΔescN as examples. The ΔsepD (T3SS competent) and ΔsepDΔescN (T3SS deficient) strains were grown in DMEM to induce type III secretion. The ΔsepDΔescN strain was grown in DMEM containing normal l-arginine (Arg) and l-lysine (Lys), whereas the ΔsepD strain was labeled by heavy isotope-labeled [13C6]Arg and [2H4]Lys. Their secreted proteins were pooled before being subjected to filtration, protein precipitation, and digestion, and LC-MS/MS analysis. B, type III secretion profiles of C. rodentium ΔlysAΔargH mutants used for SILAC analysis. The C. rodentium strains were grown in DMEM to induce type III secretion. Secreted proteins in the supernatant were analyzed in SDS-13% PAGE and stained by Coomassie G-250. Secreted proteins from an equal amount of cultures for each strain (normalized by A600) were loaded in each lane. Plasmid pCRgrlA expresses the C. rodentium LEE-encoded positive regulator GrlA.