Abstract
Shewanella oneidensis MR-1 is an important model organism for environmental research as it has an exceptional metabolic and respiratory versatility regulated by a complex regulatory network. We have developed a database to collect experimental and computational data relating to regulation of gene and protein expression, and, a visualization environment that enables integration of these data types. The regulatory information in the database includes predictions of DNA regulator binding sites, sigma factor binding sites, transcription units, operons, promoters, and RNA regulators including non-coding RNAs, riboswitches, and different types of terminators.
Availability
http://shewanella-knowledgebase.org:8080/Shewanella/gbrowserLanding.jsp
Keywords: Database, regulatory data, high temperature unfolding, Shewanella oneidensis
Background
Shewanella oneidensis MR-1 can use a diverse set of terminal electron acceptors including iron, manganese, nitrate, nitrite, fumarate, uranium, sulfur, trimethylamine-N-oxide, dimethyl sulfoxide [1] and thrive in the environments contaminated with heavy metals and radionuclides [2]. For efficient adaptation to such environments the organism has evolved a robust regulatory system [3]. A sophisticated and complex regulatory network exists to modulate the expression of genes that enable MR-1 to utilize different carbon and energy sources [4]. This regulatory information is available from both computational predictions [5–9] and experimental studies [10,11] , but its usefulness is limited because it is not available in a centralized, web accessible regulatory database.
Understanding regulation is key to gaining a systems level understanding of any living organism. Regulatory databases exist for many model organisms, including gram-positive bacterium, Bacillus subtilis, [12] and gram-negative bacterium, Escherichia coli. The regulatory information for E. coli is available in two main databases, RegulonDB [13] and EcoCyc [14]. Most of the information in these databases is manually curated. A number of eukaryotic databases [15–20] employ GBrowser from the Generic Model Organism Database Toolkit [21] to visualize the regulatory information. As a rule the type of regulators in the databases is limited, or the regulatory information is not integrated with experimentally collected data on the organism. The main objectives of the study was to develop a regulatory database for S. oneidensis MR-1 and a GBrowser based visualization environment that allows integration of the regulatory information in the analysis of diverse experimental data (experimental results from studies employing microarrays, proteomics, and/or gene mutagenesis) collected in the Shewanella knowledgebase(http://shewanellaknowledgebase.org:8080/Shewanella/).
Methodology
The information collected in the Regulatory database, which is further referred as ShewRegDB, is based on computational prediction of different regulatory elements in S. oneidensis MR-1 and published experimental data. Different Internet resources including TractorDB, RegTransBase, Rfam, RibEx, TransTermHP, PromScan, ShewCyc, ODB, MicrobesOnline, and others were used for computational predictions of the regulatory elements. Table1 (see supplementary material) summarizes number of elements located by these sources in Shewanella oneidensis MR-1. A diverse set of methods were employed by these resources/databases to find the location of the regulatory protein binding sites, operon or RNA regulators including the use of weight matrices [5], phylogenetic footprinting [5,6] building profiles or models [7,8] , motif clustering [6], clustering coexpressed genes in order to find conserved patterns in their upstream regions, comparative genomics [22], gene/domain architecture [22], decision rule based algorithm [9], combination of above approaches [5,6,22,23] or literature search by using a controlled vocabulary [24]. In addition to computational predications from the Internet resources, we also collected regulatory information based on experimental data available in literature.
The collected information was analyzed to identify basic regulatory elements (entities), their characteristics (attributes) and main types of the regulatory elements (database objects or classes) that are essential to characterize regulation in S. oneidensis. The identified database objects include DNA regulator binding sites, RNA regulators, operons and genes. At present the class of RNA regulators includes noncoding and small RNAs, different types of terminators and riboswitches. The class of DNA regulator binding sites includes binding sites of transcription and sigma factors. An entity relationship model of the database was developed according to the identified entity classes and their characteristics, then implemented as a relational database in MySQL, and populated with the collected information.
Description of the web-server
A web interface was designed to allow users to search and to download collected regulatory information. The search engine queries three main data objects of the database: DNA regulator binding sites, RNA regulators and operons. Users can search for a specific DNA regulator binding site from 4 different resources. RNA regulator search extracts information on seven types of RNA regulators of a specific gene queried by its name or locus tag. Operon search retrieves information on operon predictions from 6 different sources.
Search results for each query are displayed in the tabular format and can be downloaded as an EXCEL spreadsheet. Entries of the table have links to GBrowser and to the Shewanella knowledgebase by location of each regulatory element and by the gene locus tag. Clicking on the links provide further options on visualization of the regulatory information by GBrowser and on association of the information with the experimental data in the knowledgebase.
A visualization environment based on GBrowser [21] was developed for accessing the collected information and for its overlaying with experimental data and other genome annotations. The browser was configured for the S. oneidensis MR-1 genome and adjusted to the specificity of collected information. The data collected in the regulatory database on each database object were converted into standard gff-files for visualization of the regulatory elements on GBrowser tracks. The following visualization options are currently available:
an overview of all regulatory elements in the genome with scrolling to any selected region,
presentation of each type of element on different tracks,
different types of zooming and
displaying detailed information on each regulatory element including decorated (colored conserved regions) FASTA sequences
. The browser has four main sections:
Search, navigation, download section,
Overview,
Tracks, and
Track Selection
. Tracks in the Genome browser are organized according to main data objects in ShewRegDB and include genes, DNA regulator binding sites, sigma factor binding sites, RNA regulators, and operons (Figure 1). Two additional tracks “Experimental Data Selection” and “ShewCyc pathways” are included to provide overlay of the regulatory information with the experimental data collected in the knowledgebase and with the S. oneidensis metabolic pathway annotation from ShewCyc [29].
Figure 1.
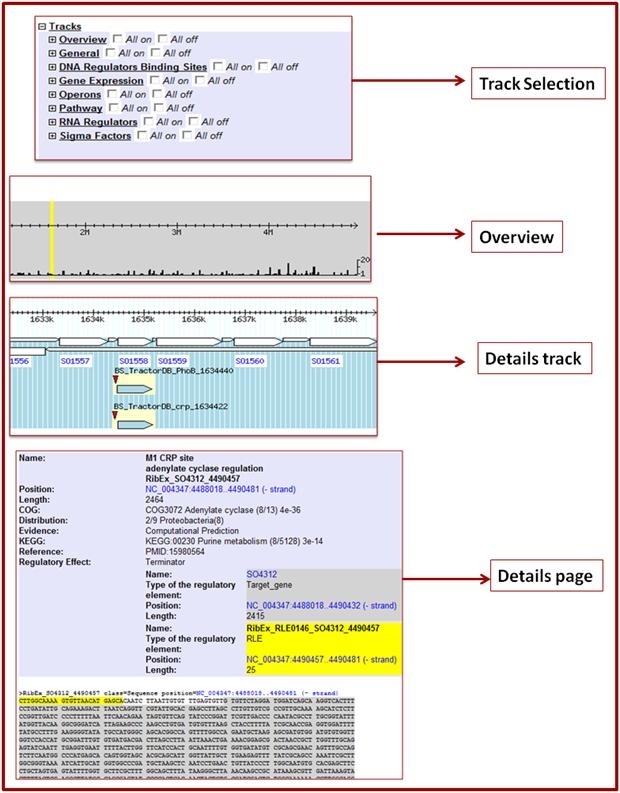
Visualization of the regulatory data from ShewRegDB in GBrowser
Utility to the biological community
ShewRegDB is unique in providing regulatory data in the framework of omics data including transcriptomics, proteomics and metabolomics datasets. This will facilitate studying the exceptional metabolic and respiratory versatility of Shewanella oneidensis MR-1, an important model organism for environmental research.
Supplementary material
Acknowledgments
Authors would like to thank Michael Galloway for system support and Loren Hauser for providing valuable comments and suggestions for this article. This research is sponsored by the U.S. Department of Energy, Office of Biological & Environmental Research. Oak Ridge National Laboratory is managed by the University of Tennessee- Battelle, LLC, for the U.S. Department of Energy under contract DEAC05- 00OR22725. The Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U. S. Department of Energy under contract DE-AC05-76RL01830.
Footnotes
Citation:Syed et al, Bioinformation 4(4): 169-172 (2009)
References
- 1.Heidelberg JF, et al. Nat Biotechnol. 2002;20:1118. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 2.Hau HH, Gralnick JA. Annu Rev Microbiol. 2007;61:237. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 3.Fredrickson JK, et al. Nat Rev Microbiol. 2008;6:592. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 4.Gralnick JA, et al. Molecular Microbiology. 2005;56:1347. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 5.González AD, et al. Nucleic Acids Res. 2005;1:D98. doi: 10.1093/nar/gki054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. Nucleic Acids Res. 2008;36:5376. doi: 10.1093/nar/gkn515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, et al. Nucleic Acids Res. 2005;1:D121. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abreu-Goodger C, Merino E. Nucleic Acids Res. 2005;1:W690. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kingsford CL, et al. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beliaev AS, et al. J Bacteriol. 2002;184:4612. doi: 10.1128/JB.184.16.4612-4616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan XF, et al. J Bacteriol. 2004;186:8385. doi: 10.1128/JB.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sierro N, et al. Nucleic Acids Res. 2008;36:D93. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gama-Castro S, et al. Nucleic Acids Res. 2008;36:D120. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keseler IM, et al. Nucleic Acids Res. 2005;1:D334. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt CJ, et al. Nucleic Acids Res. 2008;36:D719. doi: 10.1093/nar/gkm783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson RJ, et al. Nucleic Acids Res. 2008;36:D588. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swarbreck D, et al. Nucleic Acids Res. 2008;36:D1009. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreppel L, et al. Nucleic Acids Res. 2004;1:D332. doi: 10.1093/nar/gkh138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blake JA, et al. Nucleic Acids Res. 2006;1:D562. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Cruz N, et al. Nucleic Acids Res. 2005;1:D485. doi: 10.1093/nar/gki050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein LD, et al. Genome Res. 2002;12:1599. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, et al. Nucleic Acids Res. 2005;8:880. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studholme DJ, Dixon RJ. Bacteriol. 2003;185:1757. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazakov AE, et al. Nucleic Acids Res. 2007;35:D407. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, et al. PLoS Comput Biol. 2009;5:e1000338. doi: 10.1371/journal.pcbi.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda S, et al. Nucleic Acids Res. 2006;1:D358. doi: 10.1093/nar/gkj037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero PR, Karp PD. Bioinformatics. 2004;22:709. doi: 10.1093/bioinformatics/btg471. [DOI] [PubMed] [Google Scholar]
- 28.Mao F, et al. Nucleic Acids Res. 2009;37:D459. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi R, et al. Nucleic Acids Res. 2008;36:D623. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


