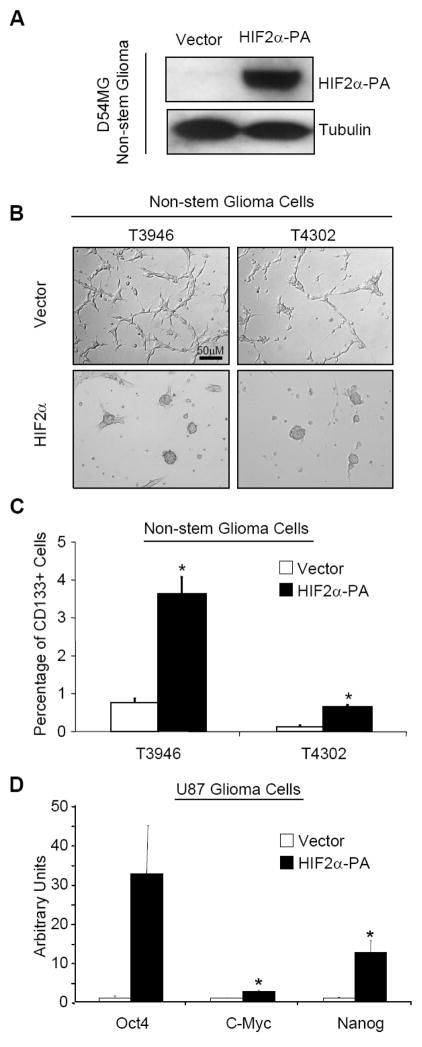
Figure 3. Ectopic expression of non-degradable HIF2α causes morphological and phenotypic changes in non-stem glioma cells.
(A) D54MG glioma cells were transfected with non-degradable HIF2α or empty pBabe vector control. Cells were allowed to recover for 3 days then lysates were harvested by Universal Lysis Buffer. Lysates were immunoblotted for the presence of HIF2α and NDRG1, a known HIF2α target. Left lane is negative control cells containing empty pBbabe vector backbone. (B) T3946 and T4302 nonstem glioma cells were transfected with non-degradable HIF2α. 2 days following transfection culture media was changed from serum containing DMEM to Neurobasal plus growth factors (detailed in Methods). Phase contrast images were taken 7 days after media change. (C) Following transfection and culture conditions outlined in (B), cells were analyzed for presence of cell surface marker CD133 (Prominin-1). White bars denote cells transfected with empty pBabe vector backbone. Fold increase was calculated by cells positive for CD133 divided by total number of viable cells gated. *, p < 0.05 with Student’s t-test comparison of cells bearing the HIF2α-PA construct against cells transfected with vector alone. Bars show standard error of the mean (SEM) for three separate measurements. (D) U87MG glioma cells transfected with non-degradable HIF2α were harvested for total RNA after puromycin selection. Semi-quantitative PCR was used to measure mRNA transcript levels of OCT4, NANOG, and c-MYC. White bars denote cells transfected with vector alone while black bars show cells transfected with HIF2α-PA. PCR data was normalized to Actin for each measurement. *, p < 0.05 with Student’s t-test comparison of cells bearing the HIF2α-PA construct against cells transfected with vector alone. Bars show standard error of the mean (SEM) for two separate reactions.