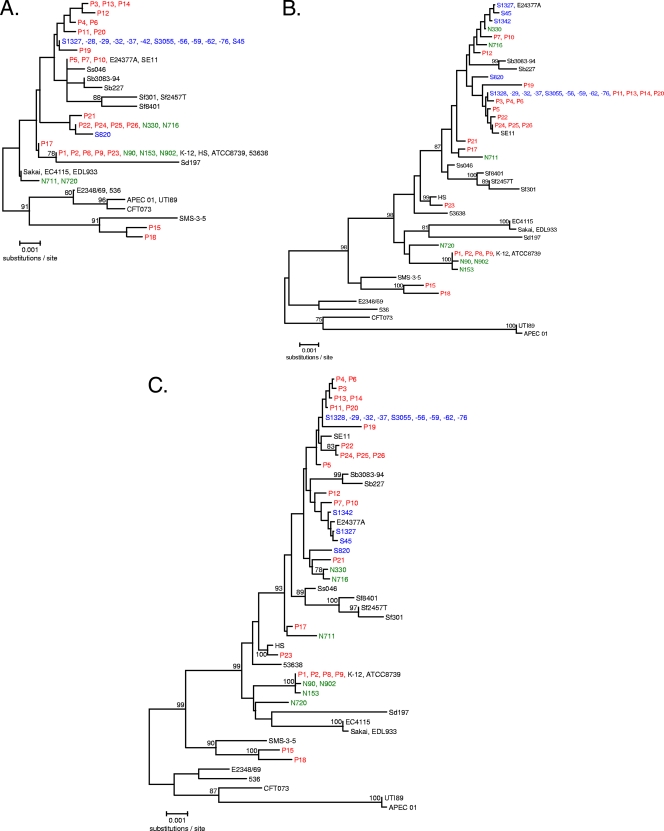
FIG. 2.
Phylogenetic relationships of acrA (A), acrB (B), and concatenated acrAB (C) alleles generated in this study and derived from genome sequences available in public sequence repositories with <3% divergence from E. coli K-12. Bootstrap values of >75 from 500 replicates are shown. N-, P-, and S-series strains are shown in green, red, and blue, respectively, while genomic sequences are in black. Genome sequences are as follows: human commensal E. coli strains ATCC 8739 (also referred to as “C”), HS, K-12, and SE11; environmental E. coli strain SMS-3-5 (from an industrial-metal-contaminated coastal environment); enterohemorrhagic E. coli (EHEC) strains EC4115, EDL933, and Sakai; enteroinvasive E. coli (EIEC) strain 53638; enteropathogenic E. coli (EPEC) strain E2348/69; ETEC strain E24377A; uropathogenic E. coli (UPEC) strains 536, CFT073, and UTI89; avian-pathogenic E. coli (APEC) strain APEC O1; Shigella boydii serotype 4 strain Sb227; S. dysenteriae serotype 1 strain Sd197; S. flexneri serotype 2a strains 2457T and 301; S. flexneri serotype 5b strain Sf8401; and S. sonnei strain Ss046.