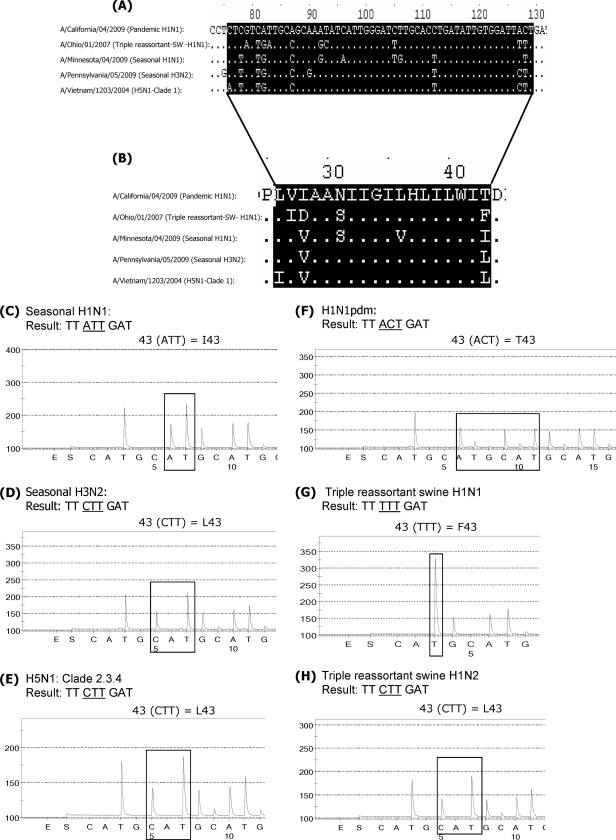
FIG. 1.
The variability in M2 protein-coding sequences among influenza A viruses can be used to differentiate between the various subtypes. (A) Partial nucleotide sequence alignment of the M2 protein-coding region from viruses of different subtypes: a pandemic virus, A/California/04/2009; a triple-reassortant swine influenza virus, A/Ohio/01/2007; seasonal H1N1 and H3N2 viruses, A/Minnesota/04/2009 and A/Pennsylvania/05/2009, respectively; and an H5N1 virus, A/Vietnam/1203/2004. (B) Partial amino acid sequence alignment of the M2 protein. Highlighted are amino acids 26 through 43. (C to H) Analysis of the sequences at residue 43. Seasonal H1N1 virus shows the presence of ATT (isoleucine) at residue 43 (I43) (C). Seasonal H3N2 virus, avian H5N1 virus, and a triple-reassortant swine H1N2 virus contain CTT (leucine) at this position, which corresponds to residue L43 (D, E, and H). Another triple-reassortant swine H1N1 virus has TTT (phenylalanine) at this position (G). The pandemic viruses have the unique sequence ACT (threonine) at residue 43 (F).