Abstract
A 3′ poly(A) tail is a common feature of picornavirus RNA genomes and the RNA genomes of many other positive-strand RNA viruses. We examined the manner in which the homopolymeric poly(A) and poly(U) portions of poliovirus (PV) positive- and negative-strand RNAs were used as reciprocal templates during RNA replication. Poly(A) sequences at the 3′ end of viral positive-strand RNA were transcribed into VPg-linked poly(U) products at the 5′ end of negative-strand RNA during PV RNA replication. Subsequently, VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNA templates were transcribed into poly(A) sequences at the 3′ ends of positive-strand RNAs. The homopolymeric poly(A) and poly(U) portions of PV RNA products of replication were heterogeneous in length and frequently longer than the corresponding homopolymeric sequences of the respective viral RNA templates. The data support a model of PV RNA replication wherein reiterative transcription of homopolymeric templates ensures the synthesis of long 3′ poly(A) tails on progeny RNA genomes.
Many positive-strand RNA viruses (e.g., members of the Picornavirales, Nidovirales, Togaviridae, Caliciviridae, and Astroviridae) have long poly(A) sequences at the 3′ termini of their RNA genomes (16, 25), yet the mechanisms by which 3′ poly(A) sequences are derived during viral replication are unclear. Picornavirus RNA genomes, including that of poliovirus (PV), have a covalently linked 5′-terminal protein called VPg (viral protein, genome linked), a 5′ untranslated region (UTR), a single large open reading frame, a 3′ UTR, and a poly(A) tail of variable length (∼20 to 150 adenosine residues) (1). RNA polymerases encoded by viruses in the order Picornavirales utilize viral proteins (and their nucleotidylylated intermediates) to prime the initiation of RNA replication at the 3′ termini of viral RNA templates (28, 29, 31, 34). In this mechanism, the viral protein VPg becomes covalently linked to the 5′ ends of both positive- and negative-strand RNAs during viral RNA replication (30, 32). PV, which is commonly studied to elucidate mechanisms of picornavirus replication, is viable when the 3′ UTR of the genome is deleted (12, 44); however, the 3′ poly(A) tail is essential for RNA replication (33, 39). The length of the 3′ poly(A) tail required for virus viability and for efficient negative-strand RNA synthesis has been examined in some detail (35, 45). PV RNAs with 3′ poly(A) tails less than 9 bases long support less than 1% of wild-type negative-strand RNA synthesis, whereas poly(A) tails ≥20 bases long support wild-type levels of negative-strand RNA synthesis (35).
In this investigation, we programmed PV RNAs with defined 3′ 84-, 51-, and 32-base poly(A) sequences [designated poly(A)(84), poly(A)(51), and poly(A)(32), respectively] into cell-free reactions that faithfully reconstitute all of the metabolic steps of viral mRNA translation (11, 22, 23) and viral RNA replication (5, 7, 27). A significant advantage of this experimental system is the ability to study one cycle of sequential negative- and positive-strand RNA synthesis (6). [α-32P]UTP and [α-32P]ATP were used to radiolabel negative- and positive-strand RNAs during PV RNA replication. The lengths of radiolabeled VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNAs and poly(A) sequences at the 3′ ends of newly synthesized positive-strand RNAs were determined by RNase T1 digestion and urea-polyacrylamide gel electrophoresis. The data revealed that VPg-linked poly(U) products were often longer than the poly(A) sequences in PV RNA templates and that long 3′ poly(A) tails on new positive-strand RNAs were synthesized during viral RNA replication. We discuss how poly(A) sequences at the 3′ end of PV RNA and VPg-linked poly(U) sequences at the 5′ end of negative-strand RNA function as reciprocal templates during viral RNA replication.
MATERIALS AND METHODS
PV cDNAs. (i) pPV A(84), pPV A(51), and pPV A(32).
The plasmid pPV A(84) was kindly provided by James B. Flanegan (University of Florida College of Medicine, Gainesville). pPV A(84) (referred to as pRNA2 in reference 13) encodes a subgenomic PV RNA replicon containing an in-frame deletion of PV nucleotides (nt) 1175 to 2956 within the capsid genes. T7 transcription of MluI-linearized pPV A(84) cDNA produces PV A(84) replicon RNA with two nonviral guanosine residues at its 5′ terminus that prevent positive-strand RNA synthesis (8, 20). While this cDNA encodes a PV RNA with a 3′ poly(A)(84) sequence, transcription of MluI-linearized pPV A(84) cDNA produced PV A(84) replicon RNAs with a distribution of 3′ poly(A) tails of ∼81 to 86 bases, as revealed by RNase T1 fingerprinting (see below).
pPV A(51) and pPV A(32) are identical to pPV A(84) except that the poly(A) sequences on the PV replicon RNAs are 51 and 32 adenines long, respectively.
(ii) pPV A(84) A79C.
pPV A(84) A79C is identical to pPV A(84) except that the 79th nucleotide in the 3′ poly(A)(84) tail was changed to cytidine. This cDNA was constructed by PCR mutagenesis using the forward primer 5′-GGACTAAAGATCCTAGGAACACTCAGG-3′ and the reverse primer 5′-TCCCCGAAAAGTGCCACCTGACGCGTTTTGTT-3′ and a pPV A(84) cDNA template. Both the PCR product and the pPV A(84) plasmid were cut with AvrII and MluI and ligated with T4 DNA ligase without gel purification.
(iii) prPV A(84).
prPV A(84), previously named pDNVR27 (29), is identical to pPV A(84) except for a 5′-terminal hammerhead ribozyme such that T7 transcription and ribozyme cleavage produce a PV ribozyme (rPV) replicon RNA possessing an authentic PV 5′ terminus to allow for both negative- and positive-strand RNA synthesis (29).
(iv) prPV A(32).
prPV A(32) is identical to prPV A(84) except that the poly(A) tail was shortened from 84 to 32 adenines.
(v) pPV VPgY3F.
pPV VPgY3F is a plasmid encoding a PV A(84) replicon RNA coding for a tyrosine-to-phenylalanine change in the third amino acid of PV protein 3B (VPg). The engineering of this mutant was described previously (9). This cDNA encoded a poly(A) tail the same length as that of pPV A(84) replicon RNA.
(vi) pPV KO CRE.
pKO CRE (referred to as pDNVR26 in reference 29) is identical to pPV A(84) except that it contains eight wobble-position mutations that disrupt the RNA cis-acting replication element (CRE) required for VPgpUpUOH synthesis without changing the amino acid sequence of PV 2CATPase (29). T7 RNA transcription of MluI-linearized pPV KO CRE cDNA produced PV CRE knockout (KO CRE) RNAs with a distribution of 3′ poly(A) tails of ∼81 to 86 residues, as revealed by RNase T1 fingerprinting (see below).
Infectious PV RNAs.
The size and sequence of the poly(A) tail of an infectious cDNA clone of PV (Mahoney type 1) with a 5′-terminal ribozyme (20) was engineered by using PCR and restriction enzymes with the following oligonucleotides: a KasI forward primer, 5′-CTCGGTGGGCGCCAAACTGCTCG 3′; an A(32) reverse primer, 5′-GCCACCTGACGCGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCTCCGAATTAAAGAAAAATTTACCCC-3′; an A(32) G10 primer, 5′-GCCACCTGACGCGTTTTTTTTTTTTTTTTTTTTTTTCTTTTTTTTTCTCCGAATTAAAGAAAAATTTACCCC-3′; an A(32) G15 primer, 5′-GCCACCTGACGCGTTTTTTTTTTTTTTTTTTCTTTTTTTTTTTTTTCTCCGAATTAAAGAAAAATTTACCCC-3′; an A(32) G20 primer, 5′-GCCACCTGA CGCGTTTTTTTTTTTTTCTTTTTTTTTTTTTTTTTTTCTCCGAATTAAAG AAAAATTTACCCC-3′; and an A(32) G25 primer, 5′-GCCACCTGACGCGTTTTTTTTCTTTTTTTTTTTTTTTTTTTTTTTTCTCCGAATTAAAGAAAAATTTACCCC-3′. KasI and MluI cleavage sites in the primers are underlined. The poly(T) regions of the reverse primers correspond to the 3′ poly(A) tail of PV RNA. KasI- and MluI-cleaved PCR products were substituted into the corresponding KasI-MluI region of the infectious cDNA clone. T7 transcription of the MluI-linearized plasmid produced T7 PV A(32) RNA. Four mutant derivatives of T7 PV A(32) RNA were engineered by introducing G substitutions into the 32-base-long poly(A) tail at positions 10, 15, 20, and 25. T7 transcription of the mutated plasmids produced T7 PV A(32) G10 RNA, T7 PV A(32) G15 RNA, T7 PV A(32) G20 RNA, and T7 PV A(32) G25 RNA.
HeLa S10 translation-replication reactions.
HeLa cell S10 extracts and HeLa cell translation initiation factors were prepared as described previously (7). HeLa S10 translation-replication reaction mixtures contained 50% (by volume) S10, 20% (by volume) translation initiation factors, 10% (by volume) 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg of creatine kinase per ml, and 155 mM HEPES-KOH [pH 7.4]), and T7 transcripts of PV replicon RNA at 45 μg/ml.
Reaction mixtures containing 2 mM guanidine HCl and the indicated PV RNA templates were incubated for 3 h at 34°C to allow for the expression of PV proteins and the formation of preinitiation RNA replication complexes (PIRCs) (6).
PV RNA replication.
PV RNA replication was assayed in reactions with PIRCs isolated from HeLa S10 translation-replication reaction mixtures. PIRCs were pelleted from HeLa S10 translation-replication reaction mixtures by centrifugation at 17,000 × g for 15 min at 4°C (26, 29). PIRCs containing the indicated PV RNA templates were resuspended in 40-μl reaction mixtures containing 27 mM HEPES-KOH [pH 7.4]; 60 mM KCH3CO2; 2.3 mM Mg(CH3CO2)2; 2.6 mM dithiothreitol; 2.3 mM KCl; 400 μg/ml creatine kinase; 30 mM creatine phosphate; 1 mM ATP; 0 to 250 μM GTP, CTP, or UTP as indicated; 50 μg/ml puromycin; and 1 μCi/μl 800-Ci/mmol [α-32P]UTP, [α-32P]GTP, [α-32P]CTP, or [α-32P]ATP as indicated. Reaction mixtures with and without 2 mM guanidine HCl and the indicated radiolabeled nucleoside triphosphate were incubated at 37°C for 1 h.
Replication complexes containing radiolabeled viral RNAs were reisolated by centrifugation at 17,000 × g for 15 min at 4°C. This allowed unincorporated radiolabel and other contaminating materials in the supernatant fractions to be discarded. Pelleted replication complexes containing radiolabeled PV RNA products of RNA replication were solubilized in 0.5% SDS buffer (0.5% sodium dodecyl sulfate, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 100 mM NaCl). Radiolabeled RNA products were phenol-chloroform extracted, ethanol precipitated, and separated by gel electrophoresis on a nondenaturing 1% agarose-1× Tris-borate-EDTA (TBE) gel.
LiCl purification of RF RNA.
Radiolabeled RNAs were resuspended in 2 M LiCl with 10 μg of an unlabeled PV replicon RNA carrier and incubated at −20°C for 1 h (3). These samples were then centrifuged at 17,000 × g for 15 min at 4°C to pellet LiCl-insoluble single-stranded RNAs and partially single-stranded RNAs. Supernatants containing double-stranded replicative form (RF) RNA were incubated in 70% ethanol at −20°C for 3 h. Double-stranded RF RNA was pelleted from 70% ethanol solutions by centrifugation at 17,000 × g for 15 min at 4°C.
RNase T1 digestion.
Radiolabeled viral RNAs were resuspended in 10-μl volumes of methylmercury hydroxide (MeHgOH) sample buffer (50 mM boric acid, 5 mM sodium borate, and 10 mM sodium sulfate). Before MeHgOH was added to each sample, a 1-μl portion of the sample was subjected to quantification by acid precipitation and scintillation counting. One microliter of 0.5 M MeHgOH was then added to the remaining 9 μl of the RNA sample, and the sample was incubated at room temperature for 10 min to denature double-stranded RNAs. MeHgOH in the sample was then inactivated by the addition of 1 μl 1.4 M β-mercaptoethanol. One microliter of RNase T1 (300 U/ml) was then added to each reaction mixture, and the mixtures were incubated at 37°C for 30 min. When indicated, proteinase K (200 μg/ml) was added to reaction mixtures, which were then incubated at 37°C for another 30 min. Reactions were terminated by the addition of a volume of 2× urea sample buffer (18 M urea, 8.9 mM Tris base, 8.9 mM boric acid [pH 8.3], 0.2 mM EDTA, 20% [wt/vol] sucrose, 0.05% [wt/vol] bromophenol blue, 0.05% xylene cyanol) equal to the volume of the reaction mixture. Radiolabeled T1 oligonucleotides in the urea sample buffer were denatured at 100°C for 3 min and separated by electrophoresis in 7 M urea-18% or 20% polyacrylamide gels in TBE buffer (89 mM Tris base, 89 mM boric acid [pH 8.3], 2 mM EDTA) for 5 h at 25 W. Radiolabeled RNAs within the gels were detected and quantified by using a phosphorimager (Bio-Rad).
3′-dCTP.
3′-dCTP was obtained from TriLink BioTechnologies (San Diego, CA).
Transfection of HeLa cells and virus quantification.
HeLa cells (∼106) in 35-mm six-well plates were transfected with T7 PV A(32), T7 PV A(32) G10, T7 PV A(32) G15, T7 PV A(32) G20, and T7 PV A(32) G25 RNAs by using TransMessenger transfection reagent according to the instructions of the manufacturer (Qiagen). Transfected HeLa cells were fed with 2 ml of cell culture medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 U per ml penicillin, and 100 μg per ml streptomycin) and incubated at 37°C. Cells were examined for cytopathic effects at 24 and 48 h posttransfection. Infectious virus in the medium was harvested following three rounds of freeze-thaw at 48 h posttransfection and quantified by a plaque assay as described previously (18).
Cloning and sequencing of poly(A) tails in PV RNA.
PV was concentrated from 6 ml of cell culture medium by centrifugation at 40,000 rpm for 2 h at 4°C using a Beckman Ti70.1 rotor. PV RNA was isolated from the concentrated virus using 4 M guanidinium thiocyanate solution (4 M guanidinium thiocyanate, 25 mM sodium citrate, 5% sodium lauroyl sarcosine, 100 mM 2-mercaptoethanol), phenol-chloroform-isoamyl alcohol, and ethanol precipitation.
The 3′ poly(A) tails of PV RNAs were converted into cDNAs, which were cloned and sequenced using methods adapted from the Andino lab (45) and the van Dyk lab (14). An RNA linker (5′-phosphate-CUACGACAGUCACCACCGAUACCCUGUACUACGCACCACG-3′) was added to the 3′ ends of PV RNAs recovered from HeLa cells or the 3′ ends of T7 transcript RNAs by using T4 single-stranded RNA ligase (New England Biolabs, Ipswich, MA). PV RNAs with the 3′ linker RNA were concentrated by ammonium acetate precipitation and transcribed to generate cDNA by using SuperScript III reverse transcriptase (Invitrogen) and a primer complementary to the 3′ end of the RNA linker (5′-CGTGGTGCGTAGTACAG-3′). cDNA corresponding to the 3′ end of PV RNA, including the poly(A) tail and RNA linker, was amplified in 30 PCR cycles with high-fidelity Phusion DNA polymerase (New England Biolabs) using a forward primer (5′-7194GGACTAAAGATCCTAGGAACACTCAGG7210-3′) corresponding to PV nt 7194 to 7210 and a reverse primer complementary to the 5′ end of the linker RNA molecule (5′-CGGTGGTGACTGTCGT-3′). PCR products were cloned using the TOPO-TA cloning kit according to the instructions of the manufacturer (Invitrogen). Chemically competent Escherichia coli TOP10 cells were transformed with the cloned products and plated onto Luria-Bertani medium containing 100 μg/ml ampicillin. Colonies were selected and screened for inserts by PCR, and plasmids were extracted using a QIAprep Spin miniprep kit (Qiagen). Plasmids were sequenced in the University of Colorado Cancer Center DNA Sequencing Core Laboratory using the above-noted forward primer corresponding to PV nt 7194 to 7210. This primer provided sequence data corresponding to the 235-base heteropolymeric sequence at the 3′ end of PV RNA, as well as the size and sequence of the poly(A) tail.
RESULTS
RNase T1 digestion and gel electrophoresis analyses of radiolabeled PV RNAs revealed the sizes of 3′ poly(A) tails in positive-strand RNA templates and the sizes of 5′ VPg-linked poly(U) sequences in negative-strand RNA products.
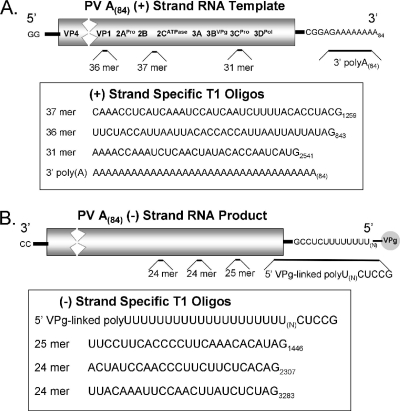
RNase T1 cleaves the 3′ end of single-stranded RNA at guanosine (G) residues and is useful in determining the lengths of homopolymeric RNA sequences in viral RNA (1). The sequences, sizes, and locations of PV RNA oligonucleotides generated by RNase T1 are predictable (Fig. 1). The largest T1 oligonucleotides within the positive strand of PV RNA include a 37-mer, a 36-mer, and a 31-mer within the heteropolymeric portion of PV RNA, as well as the 3′-terminal poly(A)(84) tail (Fig. 1A). The largest T1 oligonucleotides within the negative strand of PV RNA include the 5′-terminal VPg-linked poly(U) sequence, a 25-mer, two 24-mers (Fig. 1B), and many smaller oligonucleotides.
FIG. 1.
RNase T1 oligonucleotides in PV positive- and negative-strand RNAs. (A) RNase T1 oligonucleotides in PV A(84) positive-strand RNA. The largest RNase T1 oligonucleotides within PV A(84) positive-strand RNA are illustrated. The G residue at the 3′ end of each oligonucleotide is numbered according to its location in the PV A(84) replicon RNA used in the experiments (as described in Materials and Methods). RNase T1 digestion would liberate the 3′ poly(A) sequences from the heteropolymeric portion of PV RNA. (B) RNase T1 oligonucleotides in PV A(84) negative-strand RNA. The four largest RNase T1 oligonucleotides within PV A(84) negative-strand RNA, including their sequences, sizes, and positions relative to the 5′ end of negative-strand RNA, are illustrated.
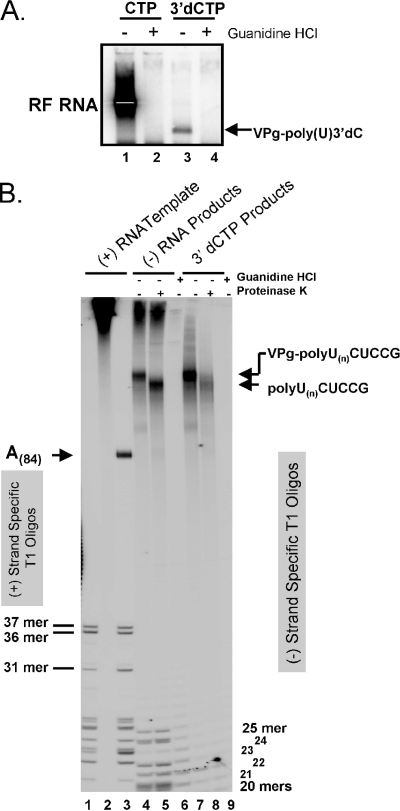
In our reactions, we can radiolabel PV RNA with α-32P-labeled nucleoside triphosphates and isolate the resulting negative-strand RNA products (Fig. 2A). Incorporation of [α-32P]ATP and [α-32P]UTP into PV RNAs can be used to radiolabel T1 oligonucleotides. Because the pattern of T1 oligonucleotides in PV positive-strand RNA is different from that in negative-strand RNA, RNase T1 can be used to distinguish PV positive-strand RNA from negative-strand RNA (6). When radiolabeled PV positive-strand RNA templates were digested with RNase T1 and the RNA fragments were separated by electrophoresis in 7 M urea-polyacrylamide gels, the predicted T1 oligonucleotides were evident (Fig. 2B, lanes 1 and 3). PV RNA radiolabeled with [α-32P]ATP and [α-32P]UTP revealed the predicted 37-mer, 36-mer, and 31-mer along with other, smaller T1 oligonucleotides from the positive-strand RNA (Fig. 2B, lanes 1 and 3). The poly(A)(84) tails at the 3′ ends of PV positive-strand RNA templates were evident only when PV RNA was radiolabeled with [α-32P]ATP and not when [α-32P]UTP was incorporated into the PV RNA (Fig. 2B, compare lanes 3 and 1). Radiolabel evident at the top of the gels corresponded to large, undigested RNA (Fig. 2B, lane 2).
FIG. 2.
VPg-linked poly(U) at the 5′ end of negative-strand RNA. (A) PV RF RNA fractionated by 1% agarose gel electrophoresis. PIRCs containing PV A(84) RNA templates were incubated in reaction mixtures containing 1 mM ATP, 250 μM GTP, 10 μM UTP, endogenous CTP from cytoplasmic extracts, and [α-32P]UTP as described in “PV RNA replication” in Materials and Methods. Guanidine HCl (2 mM) and 3′-dCTP (200 μM) were present in specific RNA replication reaction mixtures as indicated. Radiolabeled products of the reactions were separated by 1% agarose gel electrophoresis and detected by phosphorimaging. The mobilities of PV RF RNA and VPg-linked poly(U) 3′-dCMP are indicated. (B) RNase T1 oligonucleotides in PV RNAs. PV A(84) RNA templates were synthesized by T7 RNA transcription in reaction mixtures containing either [α-32P]UTP (lane 1) or [α-32P]ATP (lanes 2 and 3) as described in Materials and Methods, digested with RNase T1 (lanes 1 and 3) or untreated (lane 2), and separated by electrophoresis in 7 M urea-20% polyacrylamide (see “RNase T1 digestion” in Materials and Methods). [α-32P]UTP-radiolabeled products of PV A(84) RNA replication (lanes 4 to 9) were digested with RNase T1 and separated by electrophoresis in 7 M urea-20% polyacrylamide (see “PV RNA replication” and “RNase T1 digestion” in Materials and Methods). PV A(84) RNA products were obtained from RNA replication reaction mixtures with (lanes 6 and 9) or without (lanes 4, 5, 7, and 8) 2 mM guanidine. RNase T1 oligonucleotides were treated with proteinase K (lanes 5 and 8) or untreated (lanes 1 to 4, 6, 7, and 9). The mobilities of specific T1 oligonucleotides and VPg-linked poly(U) are indicated.
Each T1 oligonucleotide from PV RNA has a particular base composition. For example, the RNase T1 31-mer from PV positive-strand RNA templates has 6 UMP residues and 15 AMP residues (Fig. 1A). As would be predicted, the amount of radiolabel in the 31-mer was clearly greater when it was labeled with [α-32P]ATP than when it was radiolabeled with [α-32P]UTP (Fig. 2B, compare 31-mers in lanes 3 and 1). Likewise, the 3′ poly(A)(84) tail incorporated five times as much radiolabeled AMP as the 31-mer (Fig. 2B, lane 3). Thus, this method allows for detailed quantitative comparison of T1 oligonucleotides, as documented below.
VPg-linked poly(U) within the negative strand of PV RNA.
PV RNA-dependent RNA polymerase primes the initiation of negative-strand RNA synthesis with VPg and VPgpUpUOH on the 3′ poly(A) tails of input positive-strand RNA templates (40, 42). In order to determine the size of VPg-linked poly(U) at the 5′ terminus of negative-strand RNA, [α-32P]UTP was incorporated into PV negative-strand RNA as it was synthesized within PV RNA replication complexes (Fig. 2). PV positive-strand RNA templates with a 3′ poly(A)(84) sequence functioned as the template for negative-strand RNA synthesis (Fig. 2A, lane 1). Radiolabeled negative-strand RNA products remained bound to the positive-strand RNA template during the isolation of PV RNA from our reaction mixtures. Thus, negative-strand RNA migrated as double-stranded RF RNA in an agarose gel (Fig. 2A, lane 1). A 2 mM concentration of guanidine HCl, a reversible inhibitor of PV RNA replication (6), prevented the incorporation of radiolabel into PV RNA products (Fig. 2A, lanes 2 and 4). When a chain-terminating nucleotide, 3′-dCTP, was included in the reaction, RF RNA was no longer evident (Fig. 2A, lane 3); rather, a proportionally smaller amount of radiolabel was incorporated into the VPg-poly(U) product RNA that comigrated with the template in the agarose gel (Fig. 2A, lane 3). This is consistent with 3′-dCTPs being incorporated soon after transcription of the poly(A) tail into VPg-linked poly(U) (Fig. 1B). Radiolabeled VPg-poly(U) 3′-dC, which was thoroughly characterized in a recent publication from our lab (41), migrates coincidently with PV RNA templates in the agarose gel, suggesting that it is hybridized to the poly(A) tails of PV RNA templates. The bands cropped off at the very bottom of the image in Fig. 2A represent 28S rRNAs. 28S rRNAs tend to incorporate radiolabel in the experimental reactions.
RNase T1 digestion of the radiolabeled negative-strand RNA products followed by 7 M urea-polyacrylamide gel electrophoresis revealed the size of VPg-linked poly(U) at the 5′ end of negative-strand RNA (Fig. 2B, lanes 4 to 9). VPg-linked poly(U) migrated significantly more slowly than the 3′ poly(A)(84) template (Fig. 2B, compare lanes 3 and 4). To remove VPg and more precisely determine the size of poly(U), we used both proteinase K and RNase T1 digestion of VPg-linked negative-strand RNA. Proteinase K digestion increased the mobility of radiolabeled poly(U), but poly(U) was still significantly larger than the poly(A) template (Fig. 2B, compare lanes 3, 4, and 5). The heterogeneity of the sizes of radiolabeled poly(U) products was extensively characterized by experimental data presented later in this report. T1 oligonucleotides from the heteropolymeric portion of negative-strand RNA were evident near the bottom of the gel (Fig. 2B, lanes 4 and 5), and their mobility was unaffected by proteinase K digestion (Fig. 2B, compare lanes 4 and 5). RNase T1 digestion of RNA products from reaction mixtures containing 2 mM guanidine did not reveal PV RNA oligonucleotides; however, a small amount of ∼20- to 30-base-long nonviral oligonucleotides was evident (Fig. 2B, lanes 6 and 9). RNase T1 digestion of RNA products from reaction mixtures containing 3′-dCTP revealed the same VPg-linked poly(U) products as those synthesized in the absence of 3′-dCTP (Fig. 2B, lanes 7 and 8). Notably, the lanes did not contain T1 oligonucleotides from the body of full-length negative-strand RNA. Thus, 3′-dCTP prevented the synthesis of RF RNA but did not prevent the synthesis of VPg-linked poly(U) corresponding to the 5′ end of negative-strand RNA (Fig. 2).
We further validated the identity of radiolabeled VPg-linked poly(U) in the urea-polyacrylamide gels. The synthesis of radiolabeled VPg-linked poly(U) required the tyrosine hydroxyl of VPg, VPg-linked poly(U) was derived from 2 M LiCl-soluble RF RNA, and VPg-linked poly(U) was specifically radiolabeled in reactions with [α-32P]UTP in contrast to reactions with [α-32P]CTP or [α-32P]GTP (data not shown). Purification of PV RF RNA by using 2 M LiCl was advantageous because it removed significant amounts of small nonviral oligonucleotides otherwise present at the bottoms of urea-polyacrylamide gels (data not shown). Consequently, 2 M LiCl-purified RF RNA was used in subsequent experiments.
VPg-linked poly(U) sequences from VPgpUpUOH-primed and VPg-primed negative-strand RNA synthesis were indistinguishable.
As demonstrated by previously described experiments in our lab (42), both VPg and VPgpUpUOH can prime the initiation of negative-strand RNA synthesis. Therefore, we examined whether the VPg-linked poly(U) sequences from VPgpUpUOH-primed and VPg-primed negative-strand RNA synthesis were qualitatively different.
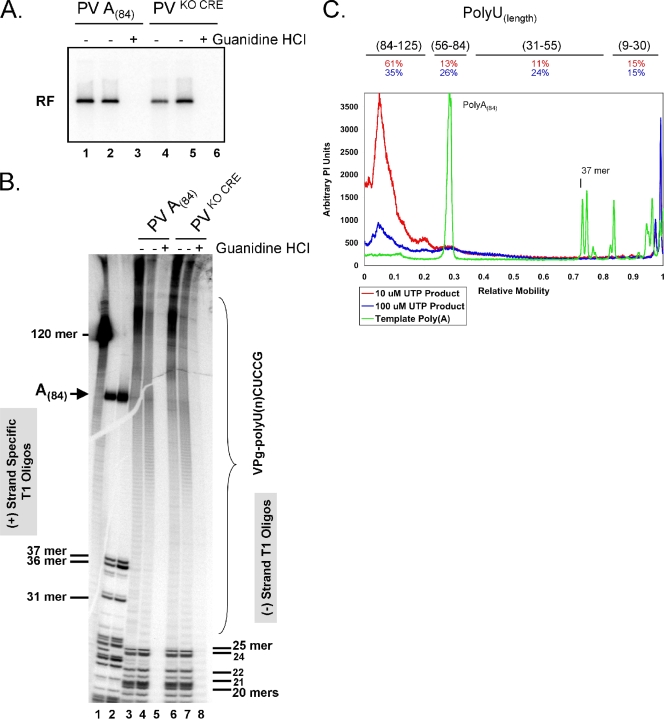
[α-32P]UTP was incorporated into PV negative-strand RNA in reactions including wild-type A(84) RNA and KO CRE RNA templates (Fig. 3A, lanes 1 to 3 versus lanes 4 to 6, respectively). KO CRE RNA templates fail to make VPgpUpUOH primers (29), and low UTP concentrations favor VPgpUpUOH priming over VPg priming (42). Consequently, we compared wild-type and KO CRE RNA templates under two reactions conditions, low and high UTP concentrations (Fig. 3). KO CRE and A(84) RNAs have identical poly(A) lengths (Fig. 3B, compare lanes 1 and 2). The 24-mer in the KO template lane is missing because the mutations used to disrupt the CRE RNA structure replaced a G residue at PV nt 4462, converting the 24-mer in wild-type RNA into a 36-mer in the KO CRE RNA (Fig. 3B, lanes 1 and 2). The T1 oligonucleotides from this region of the PV RNAs are as follows: wild-type 24-mer, 4439CAUAC UAUUA ACAAC UACAU ACAG4462, and KO CRE 36-mer, 4439CAUAC UAUUA ACAAC UAUAU CCAAU UUAAA UCCAA G4474. Mutations in the CRE RNA sequence are underlined. Note that the 24-mer in Fig. 3B, lane 1, is absent in lane 2 and that a corresponding increase in radiolabel is present in the 36-mer in lane 2. These data further highlight the reliability and quantitative sensitivity of the T1 fingerprinting technique. Guanidine HCl was used as before to inhibit PV negative-strand RNA synthesis (Fig. 3A and B). At 10 μM UTP, VPg-linked poly(U) from reaction mixtures containing VPg-primed negative-strand RNA products (Fig. 3B, lane 6) was the same length as VPg-linked poly(U) from VPgpUpUOH-primed negative-strand RNA (Fig. 3B, lane 3). Thus, VPg-linked poly(U) sequences from VPgpUpUOH-primed and VPg-primed negative-strand RNA synthesis were indistinguishable.
FIG. 3.
VPg-linked poly(U) products from wild-type and CRE-independent negative-strand RNA synthesis and the influence of UTP concentrations on the length of poly(U) sequences. (A) PV RF RNA fractionated by 1% agarose gel electrophoresis. PIRCs containing PV A(84) RNA templates (lanes 1 to 3) or PV A(84) KO CRE RNA templates (lanes 4 to 6) were incubated in reaction mixtures containing 1 mM ATP, 250 μM GTP, and 250 μM CTP, either 10 μM UTP (lanes 1, 3, 4, 6, and 7) or 100 μM UTP (lanes 2 and 5), and [α-32P]UTP, with (lanes 3 and 6) or without 2 mM guanidine HCl, as described in “PV RNA replication” in Materials and Methods. Reaction products soluble in 2 M LiCl were separated by 1% agarose gel electrophoresis and detected by phosphorimaging. The mobility of PV RF RNA is indicated. (B) RNase T1 oligonucleotides in PV RNAs. PV A(84) RNA templates (lane 1) and PV A(84) KO CRE RNA templates (lane 2) were synthesized by T7 RNA transcription in reaction mixtures containing [α-32P]ATP (see Materials and Methods), digested with RNase T1, and separated by electrophoresis in 7 M urea-20% polyacrylamide (see “RNase T1 digestion” in Materials and Methods) (lanes 1 and 2). [α-32P]UTP-radiolabeled products of PV A(84) RNA replication (lanes 3 to 5) or PV A(84) KO CRE RNA replication (lanes 6 to 8) from reaction mixtures corresponding to lanes 1 to 6 of panel A were digested with RNase T1 and separated by electrophoresis in 7 M urea-18% polyacrylamide (see “PV RNA replication” and “RNase T1 digestion” in Materials and Methods). The mobilities of specific T1 oligonucleotides and VPg-linked poly(U) are indicated. A 120-base-long RNA, corresponding to the 5′ 120 bases of PV RNA, was used as a size marker in the urea-polyacrylamide gels. (C) Size distributions of poly(A) sequences of PV A(84) RNA templates and the corresponding VPg-linked poly(U) products of RNA replication. Amounts of RNase T1 oligonucleotides from PV A(84) RNA templates (green line), VPg-linked poly(U) products from RNA replication reaction mixtures containing 10 μM UTP (red line), and VPg-linked poly(U) products from RNA replication reaction mixtures containing 100 μM UTP (blue line) are indicated. The molar amounts of VPg-linked poly(U) products were calculated based on the corresponding molar amounts of RNase T1 oligonucleotides from the heteropolymeric portion of each RNA product. PI, phosphorimaging.
[UTP] affected the length of VPg-linked poly(U).
Because UTP concentrations may affect the ability of the PV RNA polymerase to elongate efficiently during the synthesis of VPg-linked poly(U), [α-32P]UTP was incorporated into PV RF RNA in reaction mixtures containing two concentrations of UTP (10 and 100 μM) (Fig. 3). We predicted that 10 μM UTP might slow down elongation of negative-strand RNA by the PV polymerase, allowing more opportunity for nascent poly(U) RNA products to slip back onto poly(A) templates, rehybridize, and resume elongation, creating poly(U) products longer than their poly(A) templates as seen in previous figures.
We compared the lengths of VPg-poly(U) sequences synthesized in reaction mixtures containing [α-32P]UTP and either 10 or 100 μM UTP (Fig. 3B and C). Although the different UTP concentrations changed the specific activities of our reaction mixtures, equal amounts of RNase T1-digested radiolabeled negative-strand RNA were loaded into the lanes of the gel (Fig. 3B, lanes 3 and 4 and lanes 6 and 7). Proportional amounts of negative-control reaction products were loaded as well (Fig. 3B, lanes 5 and 8). Equal amounts of T1 oligonucleotides from the heteropolymeric portions of PV negative-strand RNAs were evident in the lanes (Fig. 3B, lanes 3 and 4 and lanes 6 and 7 [compare amounts of smaller T1 oligonucleotides]). VPg-linked poly(U) sequences, while heterogeneous in length under both conditions, were generally shorter on a mole-to-mole basis in reactions with 100 μM UTP than in reactions with 10 μM UTP (Fig. 3B and C). VPg-linked poly(U) ranged in length from less than 30 bases to ≥125 bases (Fig. 3B and C). In reactions including 10 μM UTP, 61% of VPg-linked poly(U) sequences were longer than the poly(A) template, whereas in reactions including 100 μM UTP, 35% of VPg-linked poly(U) sequences were longer than the poly(A) template (Fig. 3C). Increasing the [UTP] from 10 to 100 μM increased the proportion of VPg-linked poly(U) sequences 31 to 84 bases long (Fig. 3C). Thus, increased UTP concentrations modestly affected the size distribution of VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNAs.
VPg-linked poly(U) arises from internal priming along the poly(A) template rather than priming at the very 3′ end of the poly(A) tail.
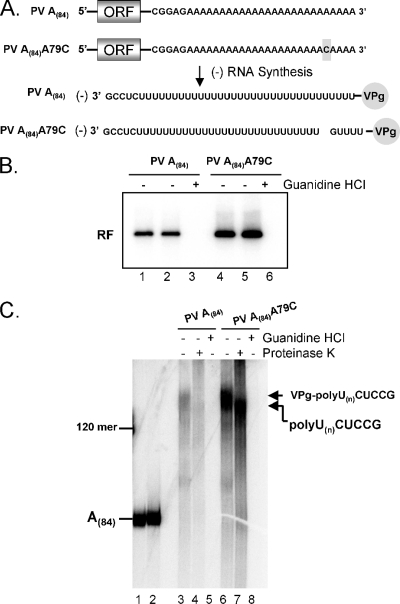
VPgpUpUOH is known to prime the initiation of positive-strand RNA synthesis at the very 3′ end of negative-strand RNA templates (34). In order to test whether VPg and/or VPgpUpUOH primes negative-strand RNA synthesis at the very 3′ end of the poly(A) template, we compared VPg-linked poly(U) synthesis from wild-type A(84) RNA templates with that from PV RNA containing a cytidine substitution for adenine 79 (A79C) within the poly(A) tail (Fig. 4A). If VPg and/or VPgpUpUOH primed the initiation of negative-strand RNA synthesis at the very 3′ end of the A79C template, a G residue would be incorporated into the VPg-linked poly(U) product ∼5 bases from the end (Fig. 4A). Furthermore, if a G was incorporated into the VPg-linked poly(U) products, RNase T1 digestion would be predicted to remove VPg and a short poly(U) sequence from the remainder of the poly(U) product (as diagrammed in Fig. 4A).
FIG. 4.
VPg-linked poly(U) products synthesized from PV A(84) and PV A(84) A79C RNA templates. (A) Diagram of PV A(84) and PV A(84) A79C RNA templates and potential products of negative-strand RNA synthesis. ORF, open reading frame. (B) PV RF RNA fractionated by 1% agarose gel electrophoresis. PIRCs containing PV A(84) RNA templates (lanes 1 to 3) or PV A(84) A79C RNA templates (lanes 4 to 6) were incubated in reaction mixtures containing 1 mM ATP, 250 μM GTP, 250 μM CTP, 10 μM UTP, 2 mM guanidine HCl (lanes 3 and 6), and [α-32P]UTP as described in “PV RNA replication” in Materials and Methods. Reaction products soluble in 2 M LiCl were separated by 1% agarose gel electrophoresis and detected by phosphorimaging. The mobility of PV RF RNA is indicated. (C) RNase T1 oligonucleotides in PV RNAs. PV A(84) and PV A(84) A79C RNA templates were synthesized by T7 RNA transcription in reaction mixtures containing [α-32P]ATP (see Materials and Methods), digested with RNase T1, and separated by electrophoresis in 7 M urea-18% polyacrylamide (see “RNase T1 digestion” in Materials and Methods) (lanes 1 and 2, respectively). [α-32P]UTP-radiolabeled products of PV A(84) RNA replication (lanes 3 to 5) or PV A(84) A79C RNA replication (lanes 6 to 8) were digested with RNase T1, treated with proteinase K (lanes 4 and 7), and separated by electrophoresis in 7 M urea-18% polyacrylamide (see “PV RNA replication” and “RNase T1 digestion” in Materials and Methods). The mobilities of specific T1 oligonucleotides and VPg-linked poly(U) products are indicated. The mobility of a 120-base-long RNA is noted to the left of the urea-polyacrylamide gel.
PV RNA containing an A79C mutation was an effective template for negative-strand RNA synthesis (Fig. 4B, lanes 4 and 5). Indeed, the amounts of negative-strand RNA from reaction mixtures containing this template were slightly larger than the amounts of negative-strand RNA from reaction mixtures containing the wild-type template in this experiment (Fig. 4B and C). The length of VPg-linked poly(U) synthesized from the PV A79C RNA was identical to that synthesized from the wild-type A(84) RNA (Fig. 4C). Furthermore, proteinase K digestion increased the mobility of A79C poly(U) products, indicating that VPg was not removed by T1 digestion from the VPg-linked poly(U) products. These results are consistent with the conclusion that VPg and/or VPgpUpUOH primed the initiation of negative-strand RNA synthesis on the poly(A) tail 5′ to the A79C mutation in the positive-strand RNA template.
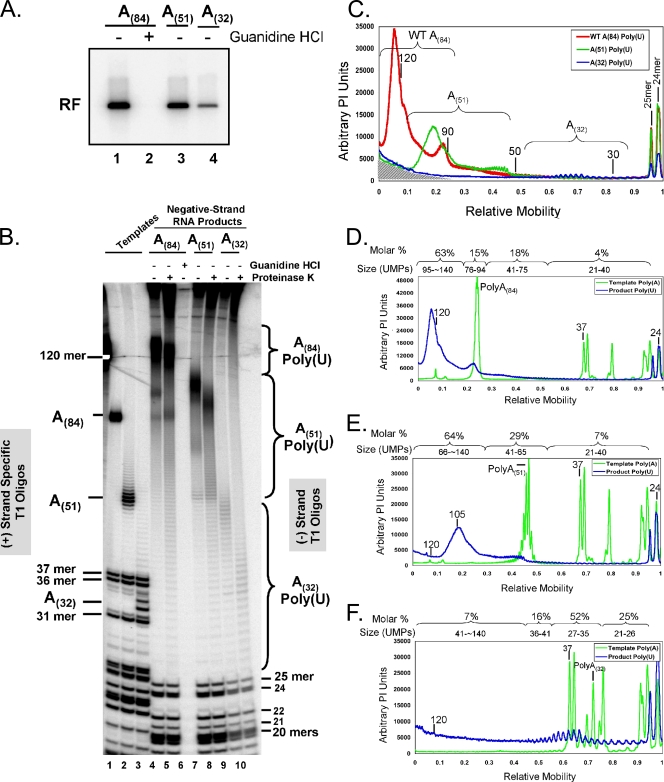
VPg-linked poly(U) from PV A(84), A(51), and A(32) RNA templates.
Because the VPg-linked poly(U) products at the 5′ ends of negative-strand RNAs made from PV poly(A)(84) RNA templates were consistently longer than the poly(A) sequence within the template, we next tested whether poly(U) length was dependent on poly(A) template length. To do this, we compared the VPg-linked poly(U) products from reaction mixtures containing templates with alternate poly(A)(84), poly(A)(51), and poly(A)(32) sequences (Fig. 5). RF RNA was evident in reaction mixtures containing PV A(84), A(51), and A(32) template RNAs (Fig. 5A, lanes 1, 3, and 4). The amounts of radiolabeled RF RNA products from reaction mixtures containing PV A(32) RNA templates were slightly smaller than those from the reaction mixtures containing PV A(84) and A(51) RNA templates (Fig. 5A). RNase T1 digestion revealed the sizes of poly(A) tails within the template RNAs (Fig. 5B, lanes 1 to 3) and the sizes of VPg-linked poly(U) sequences in negative-strand RNA products (Fig. 5B, lanes 4 to 10). Sets of T7 transcripts of A(84), A(51), and A(32) RNAs each had a small range of poly(A) sizes and were named according to the size of the poly(A) sequence in cDNA clones as explained in Materials and Methods (Fig. 5B, lanes 1 to 3, respectively). VPg-linked poly(U) from PV A(51) RNA templates was proportionately smaller than the VPg-linked poly(U) products from PV A(84) RNA templates (Fig. 5B, compare lanes 7 and 8 with lanes 4 and 5). Nonetheless, VPg-linked poly(U) products from PV A(51) RNA templates, like those from PV A(84) RNA templates, were longer than the poly(A) sequence in the templates (Fig. 5B and C). VPg-linked poly(U) products from PV A(32) RNA templates were smaller than those from either PV A(84) or A(51) RNA templates (Fig. 5B, compare lanes 9 and 10 to lanes 4 to 8). Proteinase K digestion increased the mobility of VPg-linked poly(U) products without affecting the mobility of T1 oligonucleotides from the heteropolymeric portion of negative-strand RNA (Fig. 5B, compare lanes 5, 8, and 10 to other experimental lanes).
FIG. 5.
VPg-linked poly(U) products from PV A(84), A(51), and A(32) RNA templates. (A) PV RF RNA fractionated by 1% agarose gel electrophoresis. PIRCs containing PV A(84) (lanes 1 and 2), PV A(51) (lane 3), or PV A(32) (lane 4) RNA templates were incubated in reaction mixtures containing 1 mM ATP, 250 μM GTP, 250 μM CTP, 10 μM UTP, 2 mM guanidine HCl (lane 2), and [α-32P]UTP as described in “PV RNA replication” in Materials and Methods. Reaction products soluble in 2 M LiCl were separated by 1% agarose gel electrophoresis and detected by phosphorimaging. The mobility of PV RF RNA is indicated. (B) [α-32P]ATP-labeled PV A(84) (lane 1), PV A(51) (lane 2), or PV A(32) (lane 3) RNA templates and [α-32P]UTP-labeled negative-strand RNA products of PV A(84) (lanes 4 to 6), PV A(51) (lanes 7 and 8), or PV A(32) (lanes 9 and 10) RNA replication were digested with RNase T1, untreated (lanes 1 to 4, 6, 7, and 9) or treated with proteinase K (lanes 5, 8, and 10), and separated by electrophoresis in 7 M urea-18% polyacrylamide (see Materials and Methods). The mobilities of specific T1 oligonucleotides and VPg-linked poly(U) products are indicated. The mobility of a 120-base-long RNA is noted to the left of the urea-polyacrylamide gel. (C) Size distributions of poly(U) products of RNA replication. Amounts of RNase T1 oligonucleotides from PV A(84) (red), PV A(51) (green), and PV A(32) (blue) RNA templates were determined by phosphorimager analysis of data from lanes 5, 8, and 10 of panel B. Arbitrary PI units are plotted versus the relative mobilities of products in the gel. WT, wild-type. (D) Size distributions of T1 oligonucleotides from PV A(84) RNA templates (green) and T1 oligonucleotides from the corresponding VPg-linked negative-strand RNA products (blue). Data from lanes 1 and 5 of panel B were subjected to phosphorimager analyses. Arbitrary PI units are plotted versus the relative mobilities of products in the gel. (E) Size distributions of T1 oligonucleotides from PV A(51) RNA templates (green) and T1 oligonucleotides from the corresponding VPg-linked negative-strand RNA products (blue). Data from lanes 2 and 8 of panel B were subjected to phosphorimager analyses. Arbitrary PI units were plotted versus the relative mobilities of products in the gel. (F) Size distributions of T1 oligonucleotides from PV A(32) RNA templates (green) and T1 oligonucleotides from the corresponding VPg-linked negative-strand RNA products (blue). Data from lanes 3 and 10 of panel B were subjected to phosphorimager analyses. Arbitrary PI units are plotted versus the relative mobilities of products in the gel.
The size distributions of radiolabeled poly(U) products synthesized from the different templates were determined by phosphorimaging and compared graphically (Fig. 5C; data were derived from lanes 5, 8, and 10 of Fig. 5B). There were two notable peaks of poly(U) products within the continuum of otherwise heterogeneous poly(U) sizes made from PV A(84) RNA templates: a prominent peak of poly(U) products ∼125 bases in length and a smaller peak of poly(U) products similar to the length of the A(84) templates (Fig. 5C, red line). In addition to these peaks, a continuum of poly(U) products ranged from less than 40 bases long up to the prominent peak of poly(U)(∼125) (Fig. 5B, lane 5, and C, red line). A comparable but proportionately smaller distribution of poly(U) products from the PV A(51) RNA templates was evident (Fig. 5B, lane 8, and C, green line). The amount of radiolabel incorporated into poly(U) made from PV A(32) RNA templates was smaller than that incorporated into poly(U) made from the two other templates, and the size of poly(U) made from PV A(32) RNA templates was notably smaller (Fig. 5B, lane 10, and C, blue line). The background amounts of radiolabel spilling over from undigested and partially digested RNA at the top of the gel in Fig. B are noted in the graph in Fig. 5C, where the background is indicated by diagonal lines at the bottom, corresponding predominantly to the area near the top of the gel. The peaks of radiolabel corresponding to the 25-mer and 24-mer T1 oligonucleotides were used to align T1 oligonucleotides across all lanes and are at the right-hand side of the graph (Fig. 5C, note overlapping red, green, and blue peaks for 25-mer and 24-mer oligonucleotides).
In order to compare the molar amounts of various VPg-linked poly(U) products, we calculated the molar amount of each radiolabeled poly(U) product made along the continuum and compared the size distribution to that of the poly(A) templates from which the products were synthesized (Fig. 5D, E, and F). Importantly, the molecules of VPg-poly(U) detected in the polyacrylamide gels were cumulatively present at a one-to-one molar equivalent relative to the T1 oligonucleotides from the body of the negative-strand RNA (data not shown). Of the VPg-linked poly(U) products made from PV A(84) RNA templates, 63% were longer than the poly(A) sequence in the template RNA, 15% of the VPg-linked poly(U) sequences were similar in length to the poly(A) sequence in the template, 18% were 41 to 75 bases long, and 4% were less than 40 bases long (Fig. 5D). Of the VPg-linked poly(U) products made from PV A(51) RNA templates, 64% were notably longer than the A(51) sequence in the template RNA, 29% of the VPg-linked poly(U) sequences were similar in length to the poly(A) sequence in the template, and 7% were less than 40 bases long (Fig. 5E). Only 7% of the VPg-linked poly(U) sequences made from PV A(32) RNA templates were notably longer than the A(32) sequence in the template RNA, whereas 52% of the VPg-linked poly(U) products were similar in length to the A(32) sequence in the template, 16% were slightly longer, and 25% were slightly shorter (Fig. 5F). These data indicated that the size of VPg-linked poly(U) sequences at the 5′ terminus of negative-strand RNA varied as a function of the size of the poly(A) template.
PV RNA 3′ poly(A) products of RNA replication.
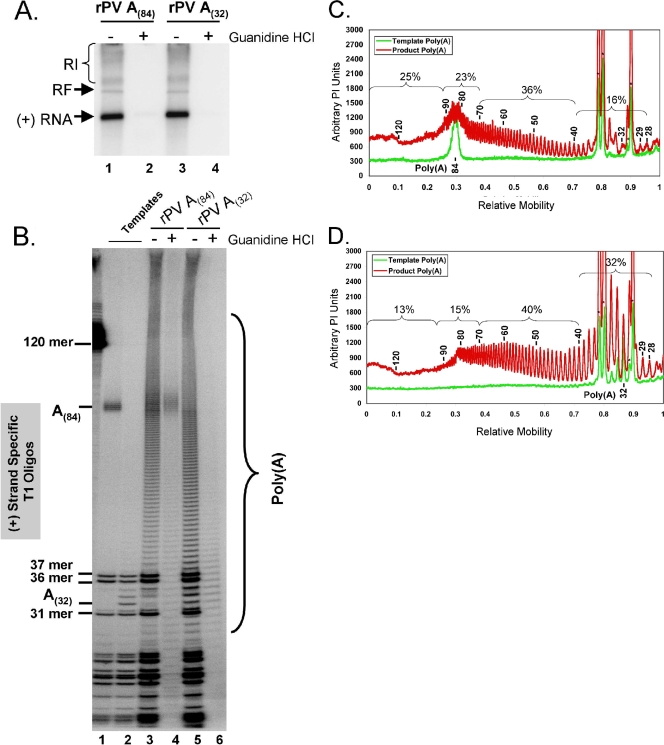
Next, we examined the lengths of poly(A) products at the 3′ ends of positive-strand RNAs made from the PV negative-strand poly(U) intermediate templates within RNA replication complexes (Fig. 6). 5′ rPV A(84) and A(32) RNA templates with authentic 5′ termini capable of synchronous and sequential negative- and positive-strand RNA synthesis were compared (Fig. 6). RF RNA and asymmetric amounts of single-stranded positive-sense RNA products were made from rPV A(84) and A(32) templates (Fig. 6A, lanes 1 and 3). These templates allowed us to determine the poly(A) lengths made at the 3′ ends of new positive-strand RNAs (Fig. 6B). A 2 mM concentration of guanidine HCl prevented the replication of rPV RNAs (Fig. 6A, lanes 2 and 4). RNase T1-digested products revealed that poly(A) sequences at the 3′ ends of newly synthesized positive-strand RNA products of PV RNA replication were very heterogeneous in length, ranging from less than 40 bases to well over 120 bases (Fig. 6B, lanes 3 and 5). The rPV A(84) poly(U) intermediate of about 125 nt templated the synthesis of 3′ poly(A) tails on new positive-strand RNAs that ranged from less than 40 bases long to over 120 bases long (Fig. 6C). Comparison to the original rPV A(84) RNA template divided the 3′ poly(A) sequences in positive-strand RNA products into four size groups: 25% were longer than the A(84) template, 23% were similar in length, 36% were somewhat shorter than the A(84) template, and 16% were less than 40 bases long (Fig. 6C). New positive-strand RNAs synthesized from rPV A(32) templates contained 3′ poly(A) sequences that were generally longer than the 32-nt poly(U) sequence in the negative-strand RNA intermediate (Fig. 6B, lane 5, and 6D). Of the 3′ poly(A) sequences in positive-strand RNA products, 28% were 70 to 140 nt long, significantly longer than the poly(A) sequence in the original rPV A(32) RNA template (Fig. 6D); 40% of the 3′ poly(A) sequences in positive-strand RNA products were 40 to 70 bases long, also longer than the poly(A) sequence in the original rPV A(32) template (Fig. 6D). Only 32% of 3′ poly(A) tail lengths were similar to that of the original rPV A(32) RNA template, at 28 to 39 bases long (Fig. 6D).
FIG. 6.
3′ poly(A) products of PV RNA replication. (A) PV RNAs fractionated by 1% agarose gel electrophoresis. PIRCs containing rPV A(84) (lanes 1 and 2) or rPV A(32) (lanes 3 and 4) RNA templates were incubated in reaction mixtures containing 1 mM ATP, 250 μM GTP, 250 μM CTP, 100 μM UTP, 2 mM guanidine HCl (lanes 2 and 4), and [α-32P]ATP as described in “PV RNA replication” in Materials and Methods. Products of the reactions were separated by 1% agarose gel electrophoresis and detected by phosphorimaging. The mobilities of PV RI, RF, and positive-strand RNAs are indicated. (B) [α-32P]ATP-labeled rPV A(84) (lane 1) or rPV A(32) (lane 2) RNA templates and [α-32P]ATP-labeled products of rPV A(84) (lanes 3 and 4) or rPV A(32) (lanes 5 and 6) RNA replication were digested with RNase T1, separated by electrophoresis in 7 M urea-18% polyacrylamide, and detected by phosphorimaging (see Materials and Methods). The mobilities of specific T1 oligonucleotides and poly(A) sequences are indicated. The mobility of a 120-base-long RNA is noted to the left of the urea-polyacrylamide gel. (C) Size distributions of T1 oligonucleotides from rPV A(84) RNA templates (green) and T1 oligonucleotides from the corresponding products of RNA replication (red). Data from lanes 1 and 3 of panel B were subjected to phosphorimager analyses. Arbitrary PI units are plotted versus the relative mobilities of products in the gel. The molar amounts of 3′ poly(A) products of RNA replication were calculated based on the corresponding molar amounts of RNase T1 oligonucleotides from the heteropolymeric portion of each positive-strand RNA product. Asterisks indicate the mobilities of heteropolymeric 37-, 36-, and 31-mers from the body of positive-strand RNA. The mobilities of rPV A(84) template and product poly(A) sequences are further annotated in the graph. (D) Size distributions of T1 oligonucleotides from rPV A(32) RNA templates (green) and T1 oligonucleotides from the corresponding products of RNA replication (red). Data from lanes 2 and 5 of panel B were subjected to phosphorimager analyses. Arbitrary PI units are plotted versus the relative mobilities of products in the gel. The molar amounts of 3′ poly(A) products of RNA replication were calculated based on the corresponding molar amounts of RNase T1 oligonucleotides from the heteropolymeric portion of each positive-strand RNA product. Asterisks indicate the mobility of heteropolymeric 37-, 36-, and 31-mers from the body of positive-strand RNA. The mobilities of rPV A(32) template and product poly(A) sequences are further annotated in the graph.
These data indicated that poly(A) sequences at the 3′ ends of positive-strand RNA products of PV RNA replication are quite heterogeneous in length. Furthermore, the poly(A) tail at the 3′ end of positive-strand RNA can be longer than the poly(A) tail and corresponding poly(U) sequence in the preceding PV RNA template (compare Fig. 6B, lane 5, with 5B, lane 10, and compare Fig. 6D with 5F).
Notably, [α-32P]ATP was found to end label the 3′ poly(A) tails of PV RNA templates in reactions including 2 mM guanidine (Fig. 6B, lanes 4 and 6). This 3′-end labeling of positive-strand template RNAs was barely detectable compared to the incorporation of [α-32P]ATP into PV RF RNA and positive-strand RNA products in reactions without guanidine (Fig. 6A, compare lanes 2 and 4 with lanes 1 and 3). Nonetheless, this [α-32P]ATP 3′-end labeling of poly(A) revealed the sizes of poly(A) sequences on PV RNA templates after several hours of incubation of the cell-free reaction mixtures (Fig. 6B, lanes 4 and 6). While [α-32P]ATP 3′-end labeling could theoretically result in modified PV RNA templates with longer poly(A) tails than those programmed into the reactions, we found that the degree of end labeling did not substantially alter the lengths of 3′ poly(A) sequences on the PV RNAs programmed into the reactions [Fig. 6B, compare template poly(A) sequences in lanes 1 and 2 with guanidine HCl negative controls in lanes 4 and 6].
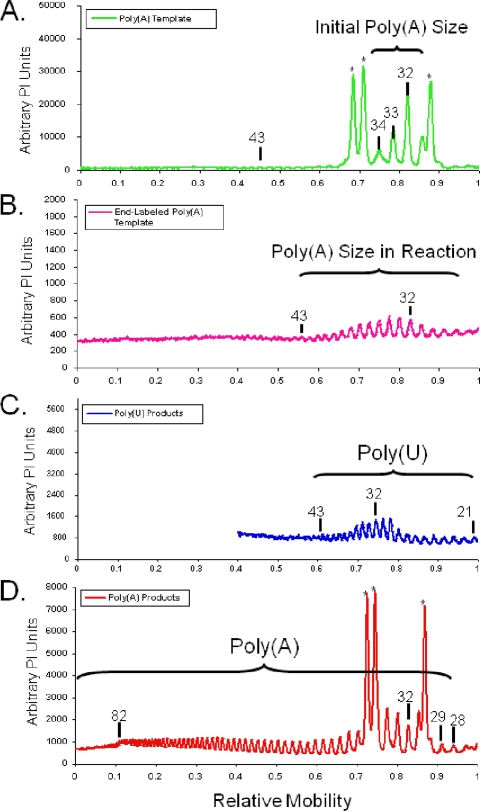
Recovery of a long 3′ poly(A) tail from a shorter 3′ poly(A) template.
The data above indicated that long 3′ poly(A) sequences were synthesized from PV RNA templates with shorter 3′ poly(A) sequences. To better illustrate this phenomenon, we graphically aligned the reciprocal templates and products of PV A(32) RNAs (Fig. 7). T7 RNA polymerase transcription of MluI-linearized cDNA coding for PV A(32) RNA produced PV RNAs with predominantly 32- to 34-base 3′ poly(A) tails (Fig. 6B, lane 2, and 7A). During the incubation of PV RNAs in HeLa S10 translation-replication reaction mixtures containing guanidine and [α-32P]ATP, the 3′ poly(A) sequences of PV RNA templates were slightly modified, as revealed by end labeling (Fig. 6B, lanes 4 and 6). 3′-end labeling revealed that the 3′ poly(A) sequences of PV A(32-34) RNA templates were slightly more heterogeneous after incubation in reaction mixtures containing cytoplasmic extracts [Fig. 7B, note 3′ poly(A) tail lengths of ∼28 to 43 bases]. The PV A(28-43) RNA templates were transcribed into 5′ VPg-linked polyU(21-43) RNA products (Fig. 7C). Despite the relatively short poly(A) sequences of and poly(U) intermediates from PV A(32) RNA templates, a fraction of newly synthesized positive-strand RNAs contained 3′ poly(A) sequences that were dramatically longer (Fig. 7D). The 3′ poly(A) sequences on positive-strand RNA products of PV A(32) RNA templates ranged from 28 to over 82 bases long (Fig. 6B, lane 5, and 7D).
FIG. 7.
rPV A(32) template sequences, poly(U) products, and corresponding poly(A) products. Size distributions of T1 oligonucleotides from [α-32P]ATP-labeled rPV A(32) RNA templates (from Fig. 4B, lane 2) (A), [α-32P]ATP end-labeled rPV A(32) RNA templates (from Fig. 4B, lane 6) (B), [α-32P]UTP-labeled PV A(32) negative-strand RNA products (from Fig. 5B, lane 10) (C), and [α-32P]ATP-labeled rPV A(32) RNA products (from Fig. 4B, lane 5) (D) are shown. PI units are plotted versus the relative mobilities of products in the gels.
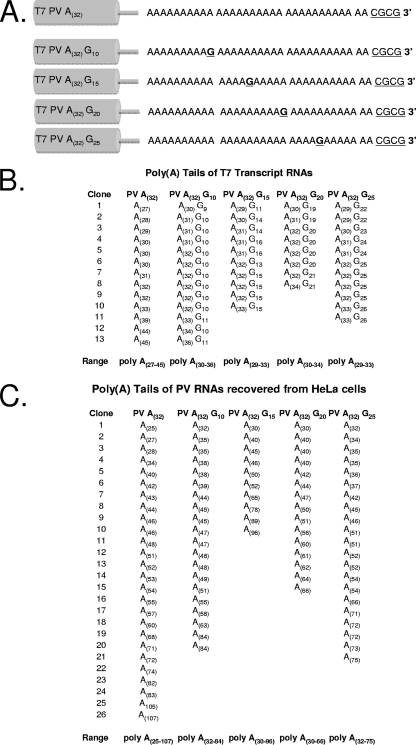
Poly(A) tails in PV RNA recovered from HeLa cells.
Next, we examined the sizes and sequences of poly(A) tails in PV RNA recovered from HeLa cells transfected with PV A(32) RNA (Fig. 8). We also tested whether G residues engineered into various positions of the 32-base-long poly(A) tail were maintained in progeny virus (Fig. 8). We expected that G residues would be maintained in progeny virus if the viral polymerase transcribed the reciprocal portions of poly(A) and poly(U) templates containing the engineered G residues. The expected 3′ termini of T7 RNA transcripts corresponding to T7 PV A(32), T7 PV A(32) G10, T7 PV A(32) G15, T7 PV A(32) G20, and T7 PV A(32) G25 RNAs are illustrated in Fig. 8A. A CGCG sequence at the very 3′ terminus of each transcript derives from the 3′ overhang of the MluI-linearized cDNA clones (Fig. 8A). The 3′ ends of T7 transcript RNAs and PV RNAs recovered from HeLa cells were TOPO-TA cloned and sequenced (Fig. 8B and C, respectively). The sizes and sequences of 3′ poly(A) tails in T7 transcript RNAs generally corresponded to the sizes and sequences of the poly(A) tails in the cDNA clones (Fig. 8B). As expected, a CGCG sequence was found at the 3′ ends of poly(A) tails in cDNA clones from T7 transcripts (data not shown). The poly(A) tails of T7 PV A(32) RNA transcripts ranged from 27 to 45 bases in length (Fig. 8B), as demonstrated by sequences from 13 independent cDNA clones. Similarly, analysis of the sequences from 8 to 13 independent cDNA clones for each construct revealed poly(A) tails of T7 PV A(32) G10, T7 PV A(32) G15, T7 PV A(32) G20, and T7 PV A(32) G25 transcript RNAs ranging from 29 to 36 bases in length (Fig. 8B). G residues were detected near the expected positions in 42 of 42 cDNA clones from PV A(32) G10, PV A(32) G15, PV A(32) G20, and PV A(32) G25 transcript RNAs (Fig. 8B). The median poly(A) tail length in the 55 cDNA clones derived from T7 transcripts was 32 bases, consistent with the cDNA templates and consistent with the data from T1 fingerprints (Fig. 5B, lane 3, and 6B, lane 2). These data indicated that the sizes and sequences of poly(A) tails in T7 transcripts of PV RNA generally corresponded to the sizes and sequences of poly(A) tails in the respective PV cDNA templates.
FIG. 8.
Poly(A) tails of PV RNAs recovered from HeLa cells. (A) Diagram of rPV A(32) RNA and mutant derivatives of rPV A(32) RNA containing G substitutions at poly(A) positions 10, 15, 20, and 25. (B) Sizes and sequences of poly(A) tails of T7 transcripts. (C) Sizes and sequences of poly(A) tails of PV RNA recovered from HeLa cells.
Cytopathic effects on cells transfected with T7 PV A(32) RNA and on cells transfected with PV A(32) G10, PV A(32) G15, PV A(32) G20, and PV A(32) G25 RNAs were evident at 24 and 48 h posttransfection. In both HeLa cells transfected with the wild type and HeLa cells transfected with mutant RNAs, ∼108 PFU of PV was produced by 48 h posttransfection. The poly(A) tails of PV RNA recovered from HeLa cells transfected with PV A(32) RNA were heterogeneous in length, ranging from 25 to 107 bases (Fig. 8C). The poly(A) tails of PV RNAs recovered from HeLa cells transfected with PV A(32) G10, PV A(32) G15, PV A(32) G20, and PV A(32) G25 RNAs were heterogeneous in length, and G residues were not detected within the poly(A) tails of recovered PV RNAs (Fig. 8C). The absence of G residues within the poly(A) tails of progeny virus from HeLa cells transfected with PV A(32) G10, PV A(32) G15, PV A(32) G20, and PV A(32) G25 RNAs indicates that the reciprocal portions of poly(A) and poly(U) templates containing the engineered G residues were not transcribed by the viral polymerase as expected or that viral RNAs containing the G substitutions within the poly(A) tail were selectively eliminated during viral RNA translation and replication in HeLa cells. The median poly(A) tail length in the 93 cDNA clones of PV RNA recovered from HeLa cells was 53 bases. The 24- to 107-base-long poly(A) tails found on PV RNAs from HeLa cells transfected with PV A(32) RNA correspond well to the lengths of poly(A) tails on progeny positive-strand RNAs formed during replication of PV A(32) RNA in PIRCs (compare data in Fig. 8C with data in Fig. 6B, lane 5, and 7D). Thus, poly(A) tails in progeny RNA were generally longer than the poly(A)(32) tails in PV RNA templates.
DISCUSSION
In this study, we exploited the synchronous and sequential replication of PV RNA within cell-free reaction mixtures (6) to examine the manner in which the homopolymeric portions of PV RNA are used as reciprocal templates. Poly(A) sequences at the 3′ end of PV RNA were transcribed into VPg-linked poly(U) products at the 5′ end of negative-strand RNA during RNA replication (Fig. 2 to 5). Subsequently, VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNA intermediates were transcribed into poly(A) sequences at the 3′ ends of progeny positive-strand RNAs (Fig. 6). RNase T1 digestion of radiolabeled PV RNAs followed by 7 M urea-20% polyacrylamide gel electrophoresis revealed the polarity of PV RNAs, the sizes of poly(A) tails within defined positive-strand RNA templates, the sizes of VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNA products, and the lengths of poly(A) tails at the 3′ ends of positive-strand RNA products of RNA replication (Fig. 2 to 7). 3′-dCTP, which prevented the elongation of PV RNA polymerase into the heteropolymeric portion of negative-strand RNA, did not prevent the synthesis of VPg-linked poly(U) (Fig. 2), consistent with the location of VPg-linked poly(U) at the 5′ end of negative-strand RNA (36, 46). Proteinase K treatment increased the mobility of radiolabeled poly(U) products, as would be expected for proteolytic removal of VPg from poly(U) products (Fig. 2, 4, and 5).
Importantly, we found that the size of VPg-linked poly(U) at the 5′ terminus of negative-strand RNA varied as a function of the size of the 3′ poly(A) sequences in PV positive-strand RNA templates (Fig. 5B and C). These data are consistent with the conclusion that poly(A) sequences at the 3′ end of positive-strand RNA function as templates for VPg-linked poly(U) synthesis at the 5′ terminus of negative-strand RNA (Fig. 5). Because VPg-linked poly(U) sequences were often longer than corresponding poly(A) templates, our data are not congruent with the conclusion that VPg-linked poly(U) sequences at the 5′ ends of picornavirus negative-strand RNAs are consistently ∼20 nucleotides long, as reported by others (45). Van Ooij et al. used SuperScript II reverse transcriptase and cDNA cloning to determine the lengths of poly(U) sequences at the 5′ ends of negative-strand RNAs (45). We found previously that SuperScript II reverse transcriptase does not efficiently elongate across long poly(U) sequences in hepatitis C virus RNA (17, 19), and van Ooij et al. did not validate the ability of their methods to detect long poly(U) sequences (45). Therefore, we suspect that van Ooij et al. failed to detect long poly(U) sequences due to technical limitations.
The presence of double-stranded RNA intermediates of replication in PV-infected HeLa cells was initially described in 1964 (2). Subsequently, the poly(A) and poly(U) sequences within replicative intermediate (RI), RF, and single-stranded positive-strand PV RNAs from PV-infected HeLa cells were characterized (15, 24, 32, 36-38, 46-49). The Baltimore and Wimmer labs purified radiolabeled RI and RF RNAs from PV-infected cells, digested the RNAs with T1 RNase and other ribonucleases, and characterized the sizes of radiolabeled poly(A) and poly(U) sequences using gradient centrifugation, urea-polyacrylamide gel electrophoresis, and nearest-neighbor analyses. Pettersson et al. found that VPg-linked poly(U) sequences at the 5′ ends of negative-strand RNAs from PV-infected HeLa cells were predominantly 120 to 140 bases in length (32), very similar to the size of VPg-linked poly(U) products made from PV A(84) RNA templates (Fig. 5B, lane 5, and C and D). Yogo and Wimmer also found long VPg-linked poly(U) sequences at the 5′ end of negative-strand RNA from RIs purified from PV-infected cells (15, 46, 47, 49). The Baltimore and Wimmer labs concluded that poly(A) at the 3′ terminus of PV RNA is genetically encoded and that the poly(A) and poly(U) portions of PV RNA templates are reciprocally transcribed by 3DPol in HeLa cells (15, 24, 36). Our data are congruent with the data and the conclusions from the Baltimore and Wimmer labs.
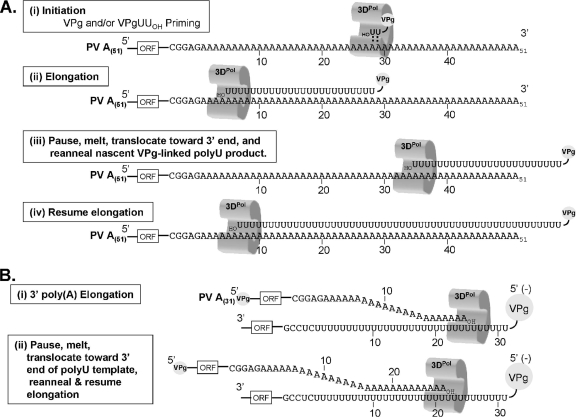
Intriguingly, a large portion of VPg-linked poly(U) products were longer than the A(84) and A(51) sequences in PV RNA templates (Fig. 5B, C, and D). In contrast, when a shorter, 3′ A(32) sequence was present in PV RNA templates, the poly(U) synthesized at the 5′ end of negative-strand RNA was almost identical in size to the poly(A) sequence in the template (Fig. 5B and C). Strikingly, 3′ poly(A) tails on new positive-strand RNAs synthesized from PV A(32) RNA templates were significantly longer than either the poly(A) sequence in the original positive-strand RNA template or the poly(U) sequences in the intermediate negative-strand RNA templates (Fig. 6 and 7). Longer poly(A) tails were also recovered from PV A(32) RNAs that had undergone replication in HeLa cells (Fig. 8). The model depicted in Fig. 9 summarizes and explains these observations. The data indicate that 3DPol and VPgpUpUOH prime the initiation of negative-strand RNA synthesis somewhere along the poly(A) tail of PV RNA (Fig. 9A, step i). Data from PV A(84) A79C templates suggest that VPgpUpUOH primes an internal site relative to the 3′ end of the poly(A) tail (Fig. 4 and Fig. 9A). 3DPol with nascent VPg-poly(U) products likely pauses (Fig. 9A, step ii) and melts, translocates toward the 3′ ends of PV RNA templates, reanneals, and resumes elongation (Fig. 9A, steps iii and iv). The same process outlined for VPg-linked poly(U) synthesis may occur during the synthesis of a poly(A) tail at the 3′ end of positive-strand RNA (Fig. 9B). An important difference during positive-strand RNA synthesis is the presence of VPg at the 5′ end of the poly(U) template (Fig. 9B) (note that VPg remains linked to the 5′ end of the negative-strand RNA template). VPg at this location may prevent 3DPol and nascent positive-strand RNA products from running off the end of the template; it may force elongating 3DPol to pause, especially on templates with shorter VPg-poly(U) sequences, such as those in rPV A(32) RNAs (Fig. 6 and 7). Reiterative transcription of poly(U)(32) templates would result in populations of newly synthesized PV RNAs with heterogeneous 3′ poly(A) tails, many of which are longer than the poly(U) sequences present within the negative-strand RNA templates [as best exemplified by the data for A(32) templates in Fig. 6, 7, and 8]. At some point, nascent positive-strand RNA molecules must dissociate from VPg-linked poly(U) templates, although at present it is unclear what regulates the cessation of poly(A) synthesis.
FIG. 9.
Reciprocal nature of poly(A) and poly(U) templates during PV RNA replication. Poly(A) sequences at the 3′ end of PV positive-strand RNA and poly(U) sequences at the 5′ end of negative-strand RNA function as reciprocal templates during PV RNA replication. (A) VPg-linked poly(U) synthesis. (B) 3′ poly(A) synthesis.
It is conceivable that poly(A) binding protein (PABP), which binds to the 3′ poly(A) tails of PV mRNAs as they translate (22, 23), may remain bound and affect the accessibility of 3′ poly(A) sequences during the initiation of negative-strand RNA synthesis. The potential contribution of PABP bound to poly(A) sequences during the initiation of negative-strand RNA synthesis has been considered previously (21, 35). Nevertheless, recent evidence suggests that picornavirus RNA replication does not require PABP (43). For this reason and for simplicity, we chose to exclude PABP from the diagrams in Fig. 9A. Nonetheless, additional experimentation is warranted to explore the potential role(s) of PABP in PV RNA replication, especially as it relates to VPg-linked poly(U) synthesis.
Our data from PIRCs are most consistent with the conclusion that the poly(A) and poly(U) portions of PV RNA templates were reciprocally transcribed by 3DPol. Nonetheless, when we engineered G residues into the poly(A) tails of PV RNA templates to test this hypothesis in HeLa cells, the G residues engineered into PV RNA templates were not recovered in the poly(A) tails of progeny virus (Fig. 8). G residues within the poly(A) tail of PV mRNA would likely disrupt normal PABP binding, potentially inhibiting viral mRNA stability, viral mRNA translation, and/or viral RNA replication. Additional experiments need to be executed to determine if G residues within the poly(A) tails interfere with particular steps of PV RNA translation and/or RNA replication.
Vesicular stomatitis virus (VSV) RNA polymerase reiteratively transcribes a short, 7-base poly(U) sequence in the intergenic region of VSV negative-strand RNA templates, leading to the synthesis of long poly(A) tails on VSV mRNA transcripts (4). The reiterative transcription of repetitive (homopolymeric) sequences by the PV RNA-dependent RNA polymerase as diagrammed in Fig. 9 may be analogous to the mechanisms used by VSV and may also be analogous to the mechanisms used by telomerases to maintain the 3′ ends of eukaryotic chromosomes (10). While the 3′ poly(A) sequences of positive-strand RNA genomes are not commonly thought of as telomeres, their repetitive sequence and location at the 3′ ends of RNA genomes is consistent with telomeric functions. Additional experimental data will be needed to determine whether the poly(A) and poly(U) portions of PV RNA templates are reciprocally transcribed (and elongated) by 3DPol and whether the viral polymerase reiteratively transcribes particular portions of the homopolymeric templates. In any case, the mechanisms appear to ensure the integrity of the 3′ end of the viral RNA genome. It will be important to further elucidate these mechanisms and determine whether similar strategies are involved in the synthesis of poly(A) tails in other polyadenylated positive-strand RNA viruses.
Acknowledgments
We thank Michelle Kelly for technical assistance.
This work was supported by NIH grants AI042189 (to D.J.B.) and T32 AI052066 (to B.P.S.).
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Ahlquist, P., and P. Kaesberg. 1979. Determination of the length distribution of poly(A) at the 3′ terminus of the virion RNAs of EMC virus, poliovirus, rhinovirus, RAV-61 and CPMV and of mouse globin mRNA. Nucleic Acids Res. 7:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore, D., Y. Becker, and J. E. Darnell. 1964. Virus-specific double-stranded RNA in poliovirus-infected cells. Science 143:1034-1036. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore, D., and M. Girard. 1966. An intermediate in the synthesis of poliovirus RNA. Proc. Natl. Acad. Sci. U. S. A. 56:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn, E. H. 1999. Telomerase, p. 609-635. In R. F. Gesteland, T. Cech, and J. F. Atkins (ed.), The RNA world: the nature of modern RNA suggests a prebiotic RNA, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Brown, B. A., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97:396-405. [DOI] [PubMed] [Google Scholar]
- 12.Brown, D. M., C. T. Cornell, G. P. Tran, J. H. Nguyen, and B. L. Semler. 2005. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J. Virol. 79:11962-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diebel, K. W., A. L. Smith, and L. F. van Dyk. 2010. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16:170-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsch-Hasler, K., Y. Yogo, and E. Wimmer. 1975. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J. Virol. 16:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, J. Q., and D. J. Barton. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J. Q., H. L. Townsend, B. K. Jha, J. M. Paranjape, R. H. Silverman, and D. J. Barton. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 81:5561-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, J. Q., G. Wroblewski, Z. Xu, R. H. Silverman, and D. J. Barton. 2004. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J. Interferon Cytokine Res. 24:664-676. [DOI] [PubMed] [Google Scholar]
- 20.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempf, B. J., and D. J. Barton. 2008. Poliovirus 2APro increases viral mRNA and polysome stability coordinately in time with cleavage of eIF4G. J. Virol. 82:5847-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf, B. J., and D. J. Barton. 2008. Poly(rC) binding proteins and the 5′ cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J. Virol. 82:5835-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen, G. R., A. J. Dorner, T. J. Harris, and E. Wimmer. 1980. The structure of poliovirus replicative form. Nucleic Acids Res. 8:1217-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall, O., P. Christian, C. M. Fauquet, A. M. King, N. J. Knowles, N. Nakashima, G. Stanway, and A. E. Gorbalenya. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Virol. 153:715-727. [DOI] [PubMed] [Google Scholar]
- 26.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 28.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. U. S. A. 74:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul, A. V., J. Yin, J. Mugavero, E. Rieder, Y. Liu, and E. Wimmer. 2003. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J. Biol. Chem. 278:43951-43960. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson, R. F., V. Ambros, and D. Baltimore. 1978. Identification of a protein linked to nascent poliovirus RNA and to the polyuridylic acid of negative-strand RNA. J. Virol. 27:357-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarnow, P. 1989. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 63:467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, N., B. J. O'Donnell, and J. B. Flanegan. 2005. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 79:3565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvestri, L. S., J. M. Parilla, B. J. Morasco, S. A. Ogram, and J. B. Flanegan. 2006. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3′ poly(A) tail. Virology 345:509-519. [DOI] [PubMed] [Google Scholar]
- 36.Spector, D. H., and D. Baltimore. 1975. Polyadenylic acid on poliovirus RNA. IV. Poly(U) in replicative intermediate and double-stranded RNA. Virology 67:498-505. [DOI] [PubMed] [Google Scholar]
- 37.Spector, D. H., and D. Baltimore. 1975. Polyadenylic acid on poliovirus RNA. II. Poly(A) on intracellular RNAs. J. Virol. 15:1418-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spector, D. H., and D. Baltimore. 1975. Polyadenylic acid on poliovirus RNA. III. In vitro addition of polyadenylic acid to poliovirus RNAs. J. Virol. 15:1432-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spector, D. H., and D. Baltimore. 1974. Requirement of 3′-terminal poly(adenylic acid) for the infectivity of poliovirus RNA. Proc. Natl. Acad. Sci. U. S. A. 71:2983-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steil, B. P., and D. J. Barton. 2009. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 139:240-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steil, B. P., and D. J. Barton. 2009. Conversion of VPg into VPgpUpUOH before and during poliovirus negative-strand RNA synthesis. J. Virol. 83:12660-12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steil, B. P., and D. J. Barton. 2008. Poliovirus cis-acting replication element-dependent VPg uridylylation lowers the Km of the initiating nucleoside triphosphate for viral RNA replication. J. Virol. 82:9400-9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svitkin, Y. V., M. Costa-Mattioli, B. Herdy, S. Perreault, and N. Sonenberg. 2007. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP). RNA 13:2330-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ooij, M. J., C. Polacek, D. H. Glaudemans, J. Kuijpers, F. J. van Kuppeveld, R. Andino, V. I. Agol, and W. J. Melchers. 2006. Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element. Nucleic Acids Res. 34:2953-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yogo, Y., M. H. Teng, and E. Wimmer. 1974. Poly(U) in poliovirus minus RNA is 5′-terminal. Biochem. Biophys. Res. Commun. 61:1101-1109. [DOI] [PubMed] [Google Scholar]
- 47.Yogo, Y., and E. Wimmer. 1973. Poly (A) and poly (U) in poliovirus double stranded RNA. Nat. New Biol. 242:171-174. [DOI] [PubMed] [Google Scholar]
- 48.Yogo, Y., and E. Wimmer. 1972. Polyadenylic acid at the 3′-terminus of poliovirus RNA. Proc. Natl. Acad. Sci. U. S. A. 69:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yogo, Y., and E. Wimmer. 1975. Sequence studies of poliovirus RNA. III. Polyuridylic acid and polyadenylic acid as components of the purified poliovirus replicative intermediate. J. Mol. Biol. 92:467-477. [DOI] [PubMed] [Google Scholar]