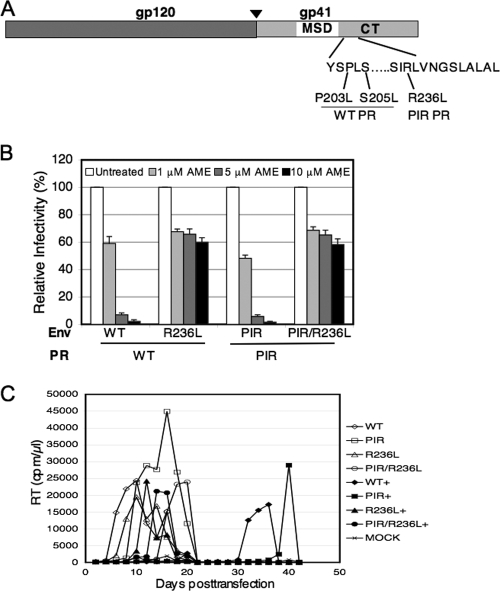
FIG. 2.
The R236L mutation confers resistance to AME in the context of WT or PIR PR. (A) Identification of the R236L mutation. The env region was PCR amplified from genomic DNA purified at the peak of viral replication from WT- and PIR-infected Jurkat cultures treated with AME, and the PCR product was sequenced as described previously (27). The organization of HIV-1 Env is indicated, with an arrowhead showing the cellular protease cleavage site between gp120 and gp41. Locations of the R236L mutation and the previously described AME-resistant mutants (P203L, S205L) selected in the context of WT PR (27, 28) are shown. MSD, membrane-spanning domain. (B) TZM-bl cells were infected with virions bearing WT or R236L Env and WT or PIR PR. The R236L mutant was constructed by QuikChange site-directed mutagenesis (Stratagene), and the PIR mutations were introduced into the pNL4-3/R236L clone as described in the legend to Fig. 1. Infections were carried out in the absence or presence of 1, 5, or 10 μM AME as described in the legend to Fig. 1. Data shown are means ± SEs of the results from three independent experiments performed in duplicate. The relative infectivities of the NL4-3/PIR, NL4-3/R236L, and NL4-3/PIR/R236L mutants, compared to that of the WT, were 97%, 87%, and 72%, respectively. (C) Five million Jurkat cells were transfected with 5 μg of the indicated molecular clones (5 μg) by using the DEAE-dextran transfection procedure and were cultured in the absence or continual presence (+) of 5 μM AME. Cultures were split 1:3 every 2 days, and supernatants were reserved for RT assay at each time point.