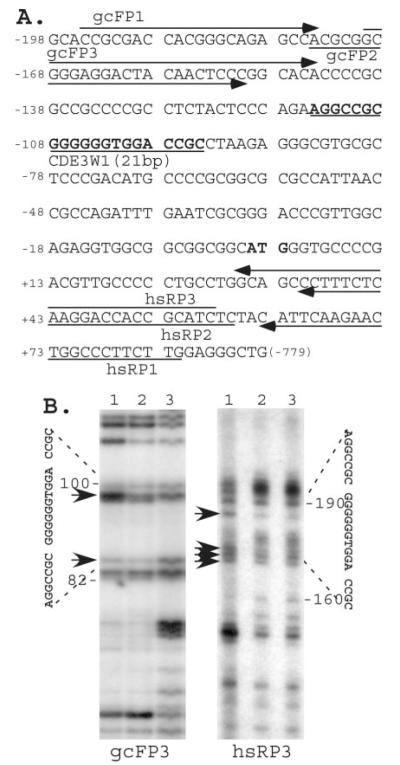
Fig. 4. The in vivo LM-PCR footprinting experiment revealed that a DNA element containing a 21-bp sequence was sensitive to in vivo DMS-mediated piperidine digestion after hedamycin treatment.
A, positions of primers used in the LM-PCR experiment are shown, and the 21-bp DNA sequence is in bold letters and underlined. B, in vivo footprinting by LM-PCR. Lane 1, arrows denote bands sensitized by hedamycin in the range of the 21-bp DNA element in the in vivo footprinting. HeLa cells were treated overnight with (lane 1) or without (lane 2) 0.2 μM hedamycin and processed by in vivo DMS-mediated piperidine digestion as described under “Experimental Procedures.” Lane 3 shows in vitro controls from direct DMS-mediated piperidine digestion of isolated untreated HeLa genomic DNA. Positions of the 21-bp sequence region and the sensitized bands from forward (gcFP3) and reverse (hsRP3) directions in the LM-PCR are indicated.