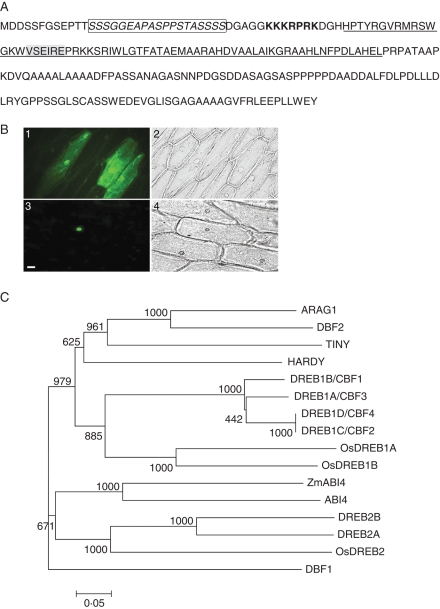
Fig. 1.
ARAG1 encodes a DREB-like protein. (A) Amino acid sequence of ARAG1. The AP2/EREBP domain is underlined. The potential nuclear localization signal, the serine/threonine-rich region and VSEIRE motif is marked with bold, italic (in box) and shading, respectively. (B) ARAG1 is a nucleus-localized protein: 1, 2 are transformants with pCAMBIA1302 vector observed in FITC filter and bright field, respectively; 3, 4 are transformants with ARAG1 over-expression construct observed in the FITC filter and bright field, respectively. Scale bar = 30 µm for 1–4. (C) Phylogenetic tree constructed by multiple sequence alignments of the AP2/EREBP domain of ARAG1 and those of other DREB proteins using Clustal W (Higgins et al., 1992) and MEGA3.1 (Kumar et al., 2004). The sequences used are: arabidopsis DREB1A/CBF3, DREB1B/CBF1, DREB1C/CBF2, DREB1D/CBF4, DREB2A, DREB2B, ABI4, HARDY and TINY (Accession nos FJ169301, FJ169278, FJ169318, NM_124578, NM_001036760, NP_187713, NP_181551 NP_181186 and NP_197953, respectively), maize DBF1, DBF2 and ZmABI4 (Accession nos AF493800, AF493799 and AY125490), rice ARAG1, OsDREB1A, OsDREB1B and OsDREB2 (Accession nos BAG98540, AF300970, AF300972, AF300971).