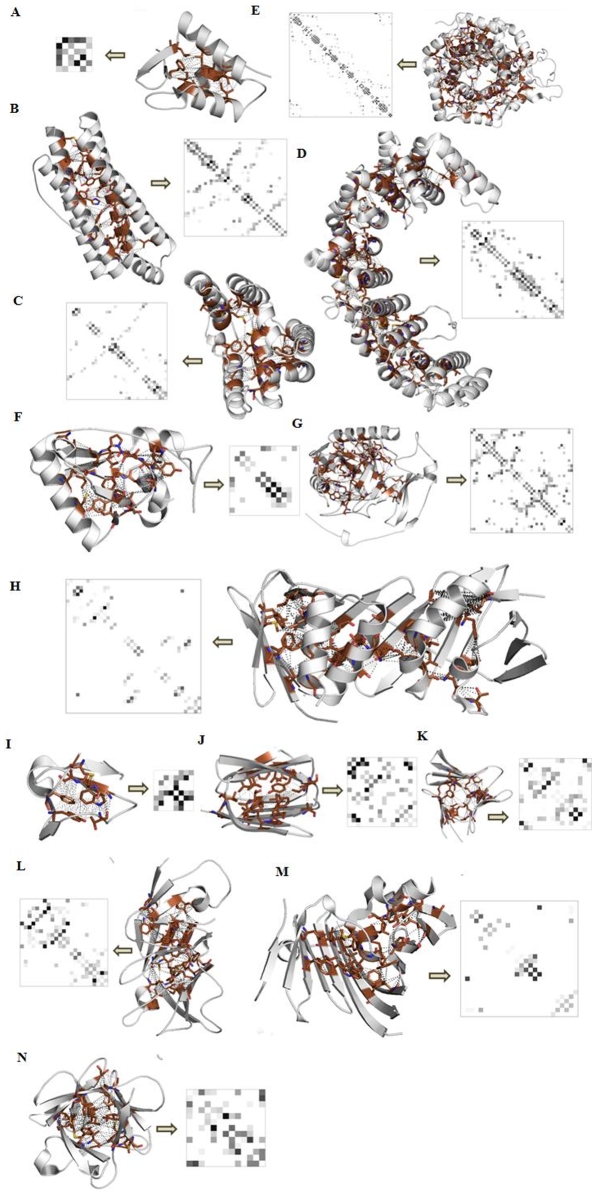
Figure 2. Snapshots from the PCAIN database used for mining fold-distinguishing signatures.
The solvent inaccessible core of domains (shaded brown) from all 1018 naturally occurring folds were identified and used to compute the PCAINs (as described in the methods section) as part of the PCAIN database. Shown herein are representative domains and PCAINs (with yellow arrow between) from the following fold families–(A.) Orthogonal α-bundle (DNA helicase RuvA subunit); (B.) Up-down α-bundle (coiled-coil); (C.) α-horseshoe (leucine-rich repeat variant); (D.) α-solenoid (peridinin-chlorophyll protein); (E.) αα-barrell (glycosyltransferase); (F.) αβ-roll (HIV reverse transcriptase); (G.) αβ-complex (cytochrome); (H.) αβ-box (proliferating cell nuclear antigen); (I.) β-ribbon (seminal fluid protein PDC-109); (J.) β-sandwich (neurophysin); (K.) β-barrel (thrombin); (L.) β-propeller (pseudo β-propeller); (M.) β-clam (outer membrane lipoprotein receptor); (N.) β-trefoil (acidic fibroblast growth factor). Fold-distinguishing PCAIN patterns observed herein motivated systemic computation of intra-fold and inter-fold correlations on a family-by-family basis, as shown in supplementary figure S5. Fold-conserved interactions are evolutionary markers and are demarcated (red stars) on the corresponding sample set of the protein family alignments in supplementary figure S3.