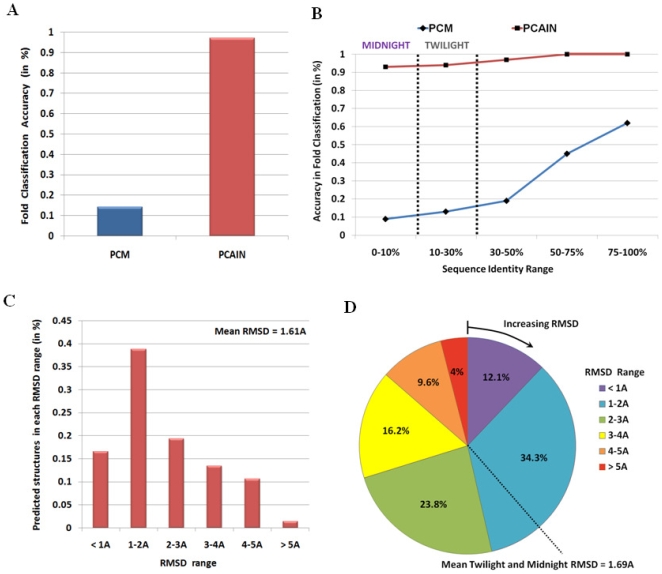
Figure 4. Applications of PCAIN as a divergence-independent metric for protein classification, anchored sequence alignment, and structure prediction.
(A.) PCAINs were computed on a general screen of unselected protein domain sequences that were not part of the database and used to accurately classify these sequences as shown, confirming the fold-specific nature of PCAINs. PCMs of these domains are seen to be ineffective as classifiers in the general sequence space. (B.) PCAIN is seen to be an effective classifier regardless of the sequence identity of the target domain towards members of its native fold and is observed to be effective even in the twilight (<30% PSI) and midnight (<10% PSI) zones. On the other hand, the PCM is observed to be highly dependent on this sequence identity and provides for some moderate classification accuracy only in the high sequence identity range. (C.) The distribution of RMSD between PCAIN-based predicted structures and the reference crystal structures for target sequences with mean RMSD of 1.61A highlights the structure prediction efficacy of the proposed method. (D.) Pie chart of RMSD distribution for test sequences in the twilight and midnight zones is shown, indicating mean RMSD of 1.69A.