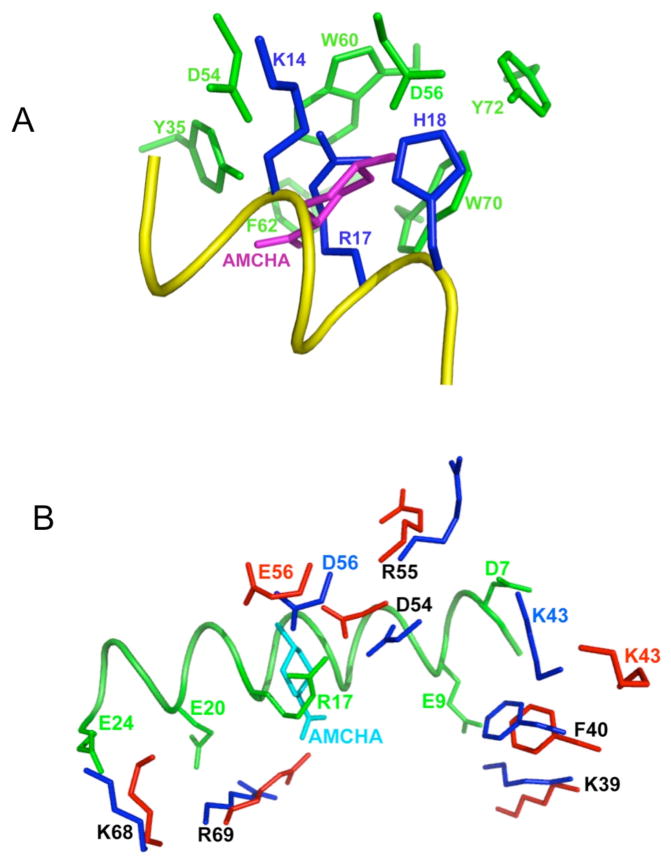
Fig. 6.
Close-up views of the intermolecular interactions comparing VEK-30/K2Pg and K2Pg/AMCHA interactions as derived from their solution structures. (A). Side-chain atoms in the hydrophobic groove (green-formed by Y35, F40, W60, W70 and Y72) and anionic center (formed by D54 and D56) of K2pg that interact with side-chain atoms from three critical amino acids of VEK-30 (blue-K14, R17, and H18) are shown. A portion of the VEK-30 helix is shown in yellow. An overlay of the K2Pg/AMCHA structure on the K2Pg/VEK-30 structure was performed and all atoms undisplayed, except for AMCHA (magenta), thus providing the relationships between the pseudo-lysine of VEK-30 and AMCHA in the K2Pg lysine binding site. (B). The binding of K2Pg to VEK-30 and K2Pg to AMCHA are overlaid for comparison and selected residues are displayed. The side-chains of K2Pg residues in the VEK-30/K2Pg complex and in the AMCHA/K2Pg complex are shown as dark blue and red sticks, respectively. The ligand, AMCHA, is colored cyan and the ribbon of VEK-30 and side-chains in VEK-30 are both colored green.