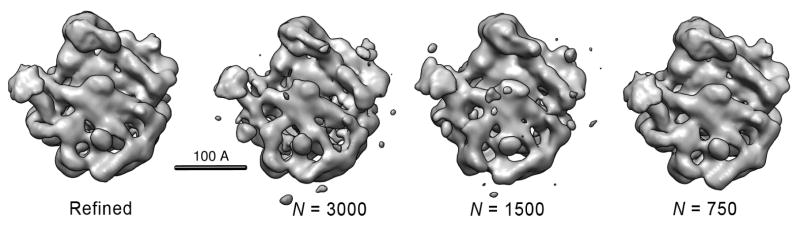
Figure 10.
Comparison of a refined model of the 50S ribosomal subunit with direct reconstructions from N = 750, 1500 and 3000 class averages. The refined model is from an Imagic reference-based alignment of the 27,121 particle data set used in this study and refined to 11.7Å resolution (3σ criterion). The remaining structures were generated directly from the voting-derived common line assignments following classification into the given numbers of input classes. The voting-based structures, for the sake of comparison, were soft masked and filtered to 15Å resolution. The structures were also flipped about the z-axis such that their handedness is consistent with the X-ray structure [Ban N. et al., 2000] and shown as the Imagic-generated 3D volumes.